The Functional Genomics group develops and utilizes state of the art mass spectrometry methods to provide metabolite and protein characterization for JBEI research. This work includes identification of novel biomass-degrading enzymes from compost samples for the Deconstruction division, characterization of the cytosolic and organelle proteomes for the Feedstocks division, and quantification of key metabolites and proteins in engineered microbes for the Biofuels and Bioproducts division. Research in the Functional Genomics Group focuses on developing methods that speed iterations of the metabolic engineering cycle (design, build, test, learn) and help characterize biomass and biomass deconstruction.
Projects
- Develop metabolomic and proteomic methods for synthetic biology applications.
- Implement quantitative analytical methods for metabolites and proteins of interest to the Feedstocks, Deconstruction, and Biofuels and Bioproducts Divisions.
- Develop a high-throughput automation pipeline for comprehensive strain engineering tasks from initial colony selection through genotyping and phenotypic analysis. ‘High-throughput, automated conversion pipeline ‘
A Skyline-based workflow for rapid development of high-throughput quantitative proteomic assays, 2016 User Group Meeting at ASMS
Starting a New Metabolic Path: JBEI and Berkeley Lab Researchers Develop Technique to Help Metabolic Engineering
-

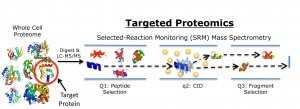
Schematic of targeted proteomics technique in which a peptide mass and a specific fragment mass are selected for SRM mass spectrometry analysis to identify and quantify a target protein. (Image from Christopher Petzold)
A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins
Featured Publications
- “Complete integration of carbene-transfer chemistry into biosynthesis,” NATURE (2023)
- “A microbial supply chain for production of the anti-cancer drug vinblastine,” NATURE (2022)
- “Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale,” NATURE COMMUNICATIONS (2022)
- “Biosynthesis of polycyclopropanated high energy biofuels,” JOULE (2022)
- “Biosynthesis and secretion of the microbial sulfated peptide RaxX and binding to the rice XA21 immune receptor,” PNAS (2019)
- “Complete biosynthesis of cannabinoids and their unnatural analogues in yeast,” NATURE (2019)
- “Lessons from Two Design-Build-Test-Learn Cycles of Dodecanol Production in Escherichia coli Aided by Machine Learning,” ACS SYNTHETIC BIOLOGY (2019)
- “Complete biosynthesis of cannabinoids and their unnatural analogues in yeast,” NATURE (2019)
- “Massively Parallel Fitness Profiling Reveals Multiple Novel Enzymes in Pseudomonas putida Lysine Metabolism,” MBIO (2019)
- “Engineering Corynebacterium glutamicum to produce the biogasoline isopentenol from plant biomass hydrolysates,” BIOTECHNOLOGY FOR BIOFUELS (2019)
- “Methyl ketone production by Pseudomonas putida is enhanced by plant-derived amino acids,” BIOTECHNOLOGY AND BIOENGINEERING (2019)
- “Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli”, Biotechnol. for Biofuels. (2018)
- “Restoration of biofuel production levels and increased tolerance under ionic liquid stress is enabled by a mutation in the essential Escherichia coli gene cydC”, Microbial Cell Factories (2018)
- “Viscous control of cellular respiration by membrane lipid composition”, Science (2018)
- “Discovery of enzymes for toluene synthesis from anoxic microbial communities”, Nature Chem. Biol. (2018)
- “Production of muconic acid in plants”, Metab. Eng. (2018)
- “Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer”, Nature Comm. (2018)
- “A bacterial pioneer produces cellulase complexes that persist through community succession”, Nature Microbiol. (2017)
- “Characterizing Strain Variation in Engineered E. coli Using a Multi-Omics-Based Workflow”, Cell Systems (2016)
- “Synthetic and systems biology for microbial production of commodity chemicals”, npj Systems Biology and Applications (2016)
- “Analytics for Metabolic Engineering”, Front. Bioeng. Biotechnol. (2015)
- “Local and global structural drivers for the photoactivation of the orange carotenoid protein”, Proc. Nat. Acad. Sci. (2015)
- “A 12 Å carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection”, Science (2015)
- “Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E.coli”, Sci. Rep. (2015)
- “The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium”, Sci. Adv. (2015)
- “Divergent Mechanistic Routes for the Formation of gem-Dimethyl Groups in the Biosynthesis of Complex Polyketides”, Angew. Chem. (2015)
- “Standard flow liquid chromatography for shotgun proteomics in bioenergy research”, Front. Bioeng. Biotechnol. (2015)
- “Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering”, Metab. Eng. (2015)
- “Correlation analysis of targeted proteins and metabolites to assess and engineer microbial isopentenol production”, Biotech. Bioeng. (2014)
- “A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins”, Metab. Eng. (2014)
- “Engineering dynamic pathway regulation using stress-response promoters”, Nat. Biotech. (2013)
- “Targeted Proteomics for Metabolic Pathway Optimization” , Methods Mol Biol. (2012)
- “Application of targeted proteomics to metabolically engineered Escherichia coli”, Proteomics (2012)
- “Modular Engineering of l-Tyrosine Production in Escherichia coli”, Appl. Environ. Microbiol. (2012)
- “Targeted Proteomics for Metabolic Pathway Optimization: Application to Terpene Production”, Metab. Eng. (2011)