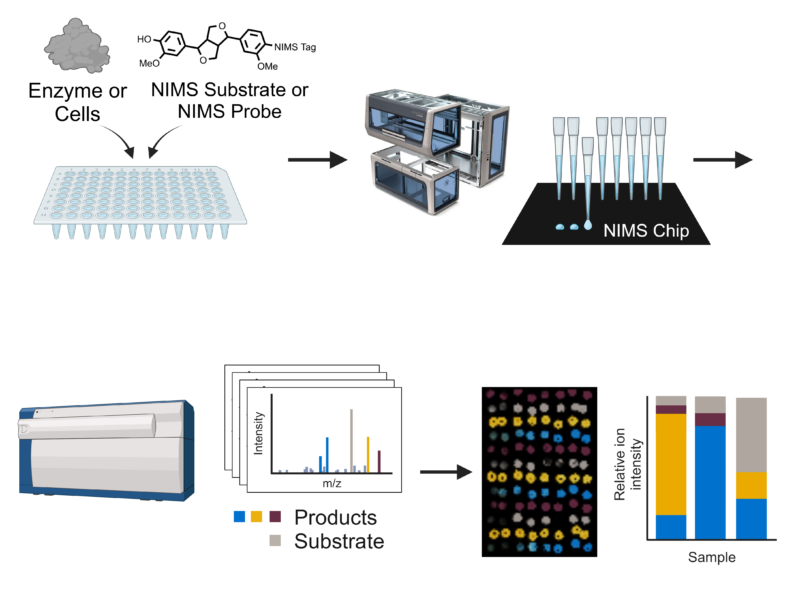
Developing cost effective methods for deconstructing plant biomass into microbial substrates and subsequent conversion to biofuels and bioproducts requires rapid analytical methodologies. To address this challenge we have developed a nanoliter-scale acoustic sample deposition and nanostructure-initiator mass spectrometry (NIMS) analysis platform to rapidly detect and characterize a wide range of enzymatic activities, but primarily focused on glyco- and lignolytic enzyme activities and substrate specificities. We are applying this approach for analysis of in vitro and cell-based expression systems to quickly screen large libraries of enzymes against a wide range of substrates including native plant biomasses. This effort will serve as the foundation in the development of this new technology that will have several applications, including enzyme “cocktail” engineering for enhanced performance in industrially relevant biorefining operating environments for the production of sugars from biomass. We have recently expanded these approaches for screening of biofuel molecules among other bioproducts. In addition, we have integrated NIMS analysis with microfluidics to create devices that enable microscale enzymatic activity analyses. Together these capabilities are enabling discovery and characterization of high performance enzymes and microbes to help advance sustainable biofuel and bioproduct platforms.
Projects
- Utilize MS Chip based assays to support the development of high performance lignocellulolytic enzymes (cellulases, hemicellulases and lignanases) screened across a matrix of conditions (T, pH, IL stability) to understand the protein features that impart ionic liquid tolerance
- Develop chip based assays to perform high throughput biofuel and bioproduct production
- Development and use of fabricated ecosystems to study bioenergy crops and their impacts with soil microbiomes
EcoFAB 3.0: Fabricated Ecosystem for Bioenergy Crops
Model plants such as Brachypodium and Arabidopsis are well-studied in controlled and sterile environments. However, there is a strong desire to extend the findings and study bioenergy crops such as sorghum. EcoFAB 3.0 is a new platform which can house a sorghum plant in a sterile controlled environment for up to 4 weeks. EcoFAB 3.0 addresses many limitations of previously used platforms (EcoFAB 2.0 and RootChip) such as ease of assembly, reusability, and lack of a dark chamber for the roots that supports gravitropism in one solution. We have successfully shown that EcoFAB 3.0 supports robust growth of sorghum that is comparable, if not better, to those grown in pots at the same age. Current studies using EcoFAB 3.0 are focussed on recreating greenhouse and field observations.

Featured Media
High Throughput Biochemistry at JBEI Using Acoustic Printing
Featured Publications
- “An engineered laccase from Fomitiporia mediterranea accelerates lignocellulose degradation” Biomolecule (2024)
- “Complete biosynthesis of QS-21 in engineered yeast” Nature (2024)
- “EcoFAB 3.0: a sterile system for studying sorghum that replicates previous field and greenhouse observations” Frontiers in Plant Science (2024)
- “Complete integration of carbene-transfer chemistry into biosynthesis” Nature (2023)
- “A combinatorial droplet microfluidic device integrated with mass spectrometry for enzyme screening” Lab Chip. (2023)
- “Mass spectrometry imaging-based assays for aminotransferase activity reveal a broad substrate spectrum for a previously uncharacterized enzyme” J. Bio. Chem. (2023)
- “Expanding extender substrate selection for unnatural polyketide biosynthesis by acyltransferase domain exchange within a modular polyketide synthase” J. Am. Soc. Chem. (2023)
- “Rapid quantification of alcohol production in microorganisms based on nanostructure-initiator mass spectrometry (NIMS)” Anal. Biochem. (2023)
- “Quantitative Analysis of the High Yield Hydrolysis of Kelp by Laminarinase and Alginate Lyase” ChemBioChem. (2023)
- “Heterologous Expression, Characterization, and Comparison of Laccases from the White Rot Causing Basidiomycete Cerrena Unicolor” Catalysis Research. (2022)
- “Faster, better and cheaper: harnessing microfluidics and mass spectrometry for biotechnology,” RSC Chem. Biol. (2021)
- “Experimental and theoretical insights into the effects of pH on catalysis of bond-cleavage by the lignin peroxidase isozyme H8 from Phanerochaete chrysosporium,” Biotechnol Biofuels (2021)
- “A multiplexed nanostructure-initiator mass spectrometry (NIMS) assay for simultaneously detecting glycosyl hydrolase and lignin modifying enzyme activities,” Sci Rep (2021)
- “A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis,” Journal of Biological Chemistry (2020)
- “A high-throughput mass spectrometric enzyme activity assay enabling the discovery of cytochrome P450 biocatalysts,”Angew Chem Int Ed Engl. (2019)
- “Lessons from two Design-Build-Test-Learn cycles of dodecanol production in Escherichia coli aided by machine learning,” ACS Synth Biol. (2019)
- “Rapid Characterization of the Activities of Lignin-Modifying Enzymes Based on Nanostructure-Initiator Mass Spectrometry (NIMS),” Biotechnology for Biofuels (2018)
- “Determination of glycoside hydrolase specificities during hydrolysis of plant cell walls using glycome profiling,” Biotechnology for Biofuels (2017)
- “OpenMSI Arrayed Analysis Toolkit: Analyzing Spatially Defined Samples Using Mass Spectrometry Imaging,” Analytical Chemistry (2017)
- “On-chip integration of droplet microfluidics and nanostructure-initiator mass spectrometry for enzyme screening”, Lab on a Chip (2017)
- “Comprehensive in Vitro Analysis of Acyltransferase Domain Exchanges in Modular Polyketide Synthases and Its Application for Short-Chain Ketone Production”, ACS Synth. Biol. (2016)
- “Comparative Community Proteomics Demonstrates the Unexpected Importance of Actinobacterial Glycoside Hydrolase Family 12 Protein for Crystalline Cellulose Hydrolysis”, mBio (2016)
- “Substrate Binding Effects of Carbohydrate Binding Modules on the Catalytic Activity of a Multifunctional Cellulase”, Biophysical Journal (2015)
- “High-Throughput Platforms for Metabolomics”, Curr Op in Chemical Biology (2015)
- “Use of Nanostructure-Initiator Mass Spectrometry to Deduce Selectivity of Reaction in Glycoside Hydrolases”, Frontiers in Bioengineering and Biotechnology (2015)
- “Development of a High Throughput Platform for Screening Glycoside Hydrolases Based on Oxime-NIMS”, Frontiers in Bioengineering and Biotechnology (2015)
- “Rapid Kinetic Characterization of Glycosyl hydrolases (GHs) based on Oxime Derivatization and Nanostructure-Initiator Mass Spectrometry (NIMS)”, ACS Chemical Biology (2014)
- “Phylogenomic Guided Identification of Industrially Relevant GH1 ?-Glucosidases Through Coupled DNA Synthesis and Nanostructure-Initiator Mass Spectrometry”, ACS Chemical Biology (2014)
- “Encoding substrates with mass tags to resolve stereospecific reactions using Nimzyme”, RCM (2012)
- “Acoustic deposition with NIMS as a high-throughput enzyme activity assay”, Analytical and Bioanalytical Chemistry (2012)
- “Colloid-based multiplexed screening for plant biomass-degrading glycoside hydrolase activities in microbial communities.”, Energy & Environmental Science (2011)
Featured Intellectual Property
- Massive Scale Enzyme Screening Using Picoliter Droplet Array on Nanostructure-Initiator Mass Spectrometry
- Microfluidic NIMS for enzyme assays
- Determining Ligninases Activities through Nanostructure Initiator Mass Spectrometry (NIMS)
- Preparation of Black Silicon Substrate for Nanostructure Initiator Mass Spectrometry (NIMS)
- NIMS-based Multiplexed Enzymatic Assays
- NIMS Skin Touch Chemical Imaging
- Rapid Discovery and Optimization of Enzyme Solutions Using Tagged Biomass and Mass Spectrometry