Research Focus
Chris Petzold is a staff scientist with Berkeley Lab’s Biological Systems and Engineering Division. He obtained his Ph. D. in Chemistry from Purdue University with emphasis in gas-phase ion chemistry and mass spectrometric methods development. After graduating from Purdue, he performed his post-doctoral work at UC-Berkeley focusing on application of mass spectrometry to glycomics and lipidomics research. Chris joined Jay Keasling’s research group in 2005 as part of the team tasked to understand the impact of metabolic engineering on the host microbe via systems biology.
As director of JBEI’s Functional Genomics Research Group, his focus is on developing mass spectrometric solutions to problems facing biofuel production. Recent work has focused on applying quantitative proteomics to metabolic engineering efforts. This work furthers rational design and optimization of novel metabolic pathways for a wide variety applications.
Projects
- Develop targeted and shotgun proteomics methods for biofuel-producing hosts
- Integrate analytical technologies with the metabolic engineering Design-Build-Test-Learn cycle
- Apply proteomic methods to help reach JBEI research goals
Featured Media
A Skyline-based workflow for rapid development of high-throughput quantitative proteomic assays, 2016 User Group Meeting at ASMS

Webinar: Jet Stream Proteomics – Enabling Standard Flow Chromatography for Robust Discovery Proteomics
Starting a New Metabolic Path: JBEI and Berkeley Lab Researchers Develop Technique to Help Metabolic Engineering
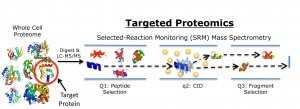
A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins
JBEI Researchers Harness Power of Microbes for Work and Winemaking
Featured Publications
- “Renewable production of high density jet fuel precursor sesquiterpenes from Escherichia coli”, Biotechnol. for Biofuels (2018)
- “Restoration of biofuel production levels and increased tolerance under ionic liquid stress is enabled by a mutation in the essential Escherichia coli gene cydC”, Microbial Cell Factories (2018)
- “Viscous control of cellular respiration by membrane lipid composition”, Science (2018)
- “Discovery of enzymes for toluene synthesis from anoxic microbial communities”, Nature Chem. Biol. (2018)
- “Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer”, Nature Comm. (2018)
- “A bacterial pioneer produces cellulase complexes that persist through community succession”, Nature Microbiol. (2017)
- “Characterizing Strain Variation in Engineered E. coli Using a Multi-Omics-Based Workflow”, Cell Systems (2016)
- “Synthetic and systems biology for microbial production of commodity chemicals”, npj Systems Biology and Applications (2016)
- “Analytics for Metabolic Engineering”, Front. Bioeng. Biotechnol. (2015)
- “Local and global structural drivers for the photoactivation of the orange carotenoid protein”, Proc. Nat. Acad. Sci. (2015)
- “A 12 Å carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection”, Science (2015)
- “Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E.coli”, Sci. Rep. (2015)
- “The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium”, Sci. Adv. (2015)
- Divergent Mechanistic Routes for the Formation of gem-Dimethyl Groups in the Biosynthesis of Complex Polyketides”, Angew. Chem. (2015)
- “Standard flow liquid chromatography for shotgun proteomics in bioenergy research”, Front. Bioeng. Biotechnol. (2015)
- “Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering”, Metab. Eng. (2015)
- “Correlation analysis of targeted proteins and metabolites to assess and engineer microbial isopentenol production”, Biotech. Bioeng. (2014)
- “A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins”, Metab. Eng. (2014)
- “Engineering dynamic pathway regulation using stress-response promoters”, Nat. Biotech. (2013)
- “Targeted Proteomics for Metabolic Pathway Optimization”, Methods Mol Biol. (2012)
- “Application of targeted proteomics to metabolically engineered Escherichia coli”, Proteomics (2012)
- “Modular Engineering of l-Tyrosine Production in Escherichia coli”, Appl. Environ. Microbiol. (2012)
- “Targeted Proteomics for Metabolic Pathway Optimization: Application to Terpene Production”, Metab. Eng. (2011)