Software engineers, research associates, and automation engineers in the Synthetic Biology Informatics Group develop and demonstrate wetware, software, and laboratory automation devices that facilitate, accelerate, and standardize the engineering of microbes that produce fuels and chemicals from cellulosic biomass.
Projects
- ICE Repository of microbial strains, (DNA and protein) sequences, and plant seeds/lines
- Bioparts.org Search portal across repositories of strains and sequences (ICE and beyond)
- EDD Experiment Data Depot
Featured Media
First Author Conversations Podcast. Episode 3: Application of Synthetic Biology

Understanding and Designing Flanking Homology DNA Assembly Experiments
Nathan Hillson talks with the Risk Group about the: “BioEconomy: Promise and Perils of Synthetic Biology”
Nathan Hillson talks about the role of software development at JBEI
Microfluidic Device Mixes And Matches DNA For Synthetic Biology

Cold Spring Harbor Laboratory Seminar: Synthetic Biology Scalability and Responsible Innovation

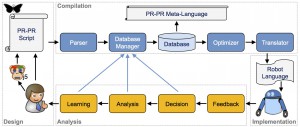
Training Your Robot the PR-PR Way
DNA Construction Software Saves Time, Resources and Money
Featured Publications
- “Machine learning-led semi-automated medium optimization reveals salt as key for flaviolin production in Pseudomonas putida” Commun Biol (2025)
- “Binary vector copy number engineering improves Agrobacterium-mediated transformation“. Nat Biotechnol (2024)
- “Automated Platform for the Plasmid Construction Process” ACS Synthetic Biology (2023)
- “Alkaline-SDS cell lysis of microbes with acetone protein precipitation for proteomic sample preparation in 96-well plate format” PLOS ONE (2023)
- “Ten simple rules for managing laboratory information.” PLoS computational biology (2023)
- “Artificial Intelligence and Machine Learning for Bioenergy Research: Opportunities and
Challenges” U.S. Department of Energy Office of Science and Office of Energy Efficiency and Renewable Energy (2023) - “Perspectives for self-driving labs in synthetic biology” Current Opinion in Biotechnology (2023)
- “Scalable and automated CRISPR-based strain engineering using droplet microfluidics” Microsyst Nanoeng (2022)
- “Modular automated bottom-up proteomic sample preparation for high-throughput Applications”. PLoS ONE (2022)
- “Making Security Viral: Shifting Engineering Biology Culture and Publishing”. ACS Synthetic Biology (2022)
- “BioParts—A Biological Parts Search Portal and Updates to the ICE Parts Registry Software Platform” ACS Synthetic Biology (2021)
- “Multiomics Data Collection, Visualization, and Utilization for Guiding Metabolic Engineering“. Frontiers in Bioengineering and Biotechnology (2021)
- “Machine learning for metabolic engineering: A review” Metabolic Engineering (2020)
- “Repurposing a microfluidic formulation device for automated DNA construction” PLOS One (2020)
- “An Integrated Computer-Aided Design and Manufacturing Workflow for Synthetic Biology” DNA Cloning and Assembly (2020)
- “Adenosine Triphosphate and Carbon Efficient Route to Second Generation Biofuel Isopentanol” ACS Synthetic Biology (2020)
- “Fluorescent amplification for next generation sequencing (FA-NGS) library preparation” BMC Genomics (2020)
- “Enhancing Terminal Deoxynucleotidyl Transferase Activity on Substrates with 3′ Terminal Structures for Enzymatic De Novo DNA Synthesis” Genes (2020)
- “Mevalonate Pathway Promiscuity Enables Noncanonical Terpene Production” ACS Synthetic Biology (2019)
- “An automated ‘cells-to-peptides’ sample preparation workflow for high-throughput, quantitative proteomic assays of microbes” J Proteome Res (2019)
- “Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases” J Ind Microbiol Biotechnol (2019)
- “Lessons from Two Design–Build–Test–Learn Cycles of Dodecanol Production in Escherichia coli Aided by Machine Learning” ACS Synthetic Biology (2019)
- “New Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases” JIMB (2019)
- “Building a global alliance of biofoundries“, Nature Communications (2019)
- “Parallel Integration and Chromosomal Expansion of Metabolic Pathways“, ACS Synthetic Biology (2018)
- “Jungle Express is a versatile repressor system for tight transcriptional control“, Nature Communications (2018)
- “De novo DNA synthesis using polymerase-nucleotide conjugates“, Nature BioTech (2018)
- “Biochemical Characterization of β-Amino Acid Incorporation in Fluvirucin B2 Biosynthesis“, ChemBioChem (2018)
- “Isolation and characterization of novel mutations in the pSC101 origin that increase copy number”, Scientific Reports (2018)
- “A combinatorial approach to synthetic transcription factor-promoter combinations for yeast strain engineering” Yeast (2018)
- “Oxidative cyclization of prodigiosin by an alkylglycerol monooxygenase-like enzyme”, Nature Chemical Biology (2017)
- “The Experiment Data Depot: a web-based software tool for biological experimental data storage, sharing, and visualization”, ACS Synthetic Biology (2017)
- “Droplet Microfluidics for Synthetic Biology”, Lab on a Chip (2017)
- “Streamlining the Design-to-Build transition with Build-Optimization Software Tools (BOOST)”, ACS Synthetic Biology (2016)
- “A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae”, Nucleic Acids Research (2016)
- “Develop an improved metabolic engineering method for modifying microorganisms for biofuel production from cellulosic sugars”, JBEI end-of-year report (2016)
- “New tools available for facilitating the design of engineered pathways in microorganisms for biofuel production from cellulosic sugars”, JBEI quarterly report (2016)
- “Improving Synthetic Biology Communication: Recommended Practices for Visual Depiction and Digital Submission of Genetic Designs”, ACS Synthetic Biology (2016)
- “Investigation of Proposed Ladderane Biosynthetic Genes from Anammox Bacteria by Heterologous Expression in E. coli“, PLOS ONE (2016)
- “End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis”, J Biol Eng (2016)
- “A Droplet Microfluidic Platform for Automating Genetic Engineering”, ACS Synthetic Biology (2016)
- “SBOL Visual: A Graphical Language for Genetic Designs”, PLOS Biology (2015)
- “A Versatile Microfluidic Device for Automating Synthetic Biology”, ACS Synthetic Biology (2015)
- “Industrialization of Biology: A Roadmap to Accelerate the Advanced Manufacturing of Chemicals“, The National Academies Press (2015)
- “The Joint Genome Institute’s synthetic biology internal review process”, Journal of Responsible Innovation (2015)
- “Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering”, Metabolic Engineering (2014)
- “Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models”, PLOS ONE (2014)
- “The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology”, Nat Biotechnol (2014)
- “The Plant Glycosyltransferase Clone Collection for Functional Genomics”, Plant J. (2014)
- “Cloud-Enabled Microscopy and Droplet Microfluidic Platform for Specific Detection of Escherichia coli in Water”, PLoS ONE (2014)
- “PR-PR Cross-Platform Laboratory Automation System”, ACS Synthetic Biology (2014)
- “j5 DNA Assembly Design Automation”, DNA Cloning and Assembly Methods (2014)
- “Development of a broad-host synthetic biology toolbox for ralstonia eutropha and its application to engineering hydrocarbon biofuel production”, Microbial Cell Factories (2013)
- “The Fusion of Biology, Computer Science, and Engineering – towards efficient and successful synthetic biology”, Perspectives in Biology and Medicine (2013)
- “PaR-PaR Laboratory Automation platform”, ACS Synthetic Biology (2012)
- “Design, Implementation and Practice of JBEI-ICE: An Open Source Biological Part Registry Platform and Tools”, Nucleic Acids Research (2012)
- “DeviceEditor visual biological CAD canvas”, Journal of Biological Engineering (2012)
- “j5 DNA Assembly Design Automation Software”, ACS Synthetic Biology (2012)
- “DNA Assembly Method Standardization for Synthetic Biomolecular Circuits and Systems”, Design and Analysis of Bio-molecular Circuits (2011)
Intellectual Property
- Scar-Less Multi-Part DNA Assembly Design Automation. (2019) U.S. Patent 11,749,379
- Scar-Less Multi-Part DNA Assembly Design Automation. (2016) U.S. Patent 10,373,703
- Scar-Less Multi-Part DNA Assembly Design Automation. (2012) U.S. Patent 9,361,427
- Microfluidic Platform for Synthetic Biology Applications. (2012) U.S. Patent Application US20120258487.