Genome-resolved metagenomics has enabled a detailed view of microbial communities adapted to grow on biomass substrates. This view has identified new capabilities of cellulases and identified uncultivated bacteria involved in biomass deconstruction.
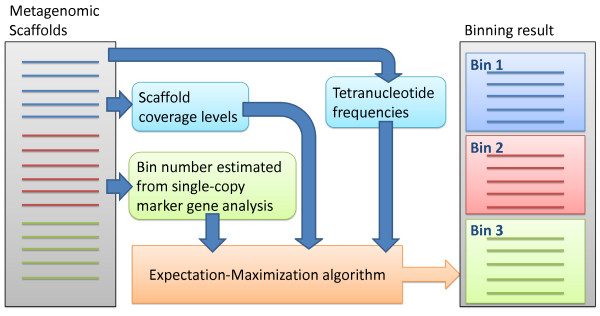
MaxBin: Automated Sorting Through Metagenomes
Reconstructing genomes from metagenomics data provides a powerful way to understand what processes and interactions toccur in microbial communities. MaxBin is an automated algorithm to reconstruct genomes from assembled metagenomic data. In this paper in Microbiome by Wu et al. (doi: 10.1186/2049-2618-2-26), a description of the MaxBin algorithm was presented along with a reconstruction of genomes from two cellulolytic consortia (consortium 1 and consortium 2) that identified a new cellulolytic myxobacterial species.
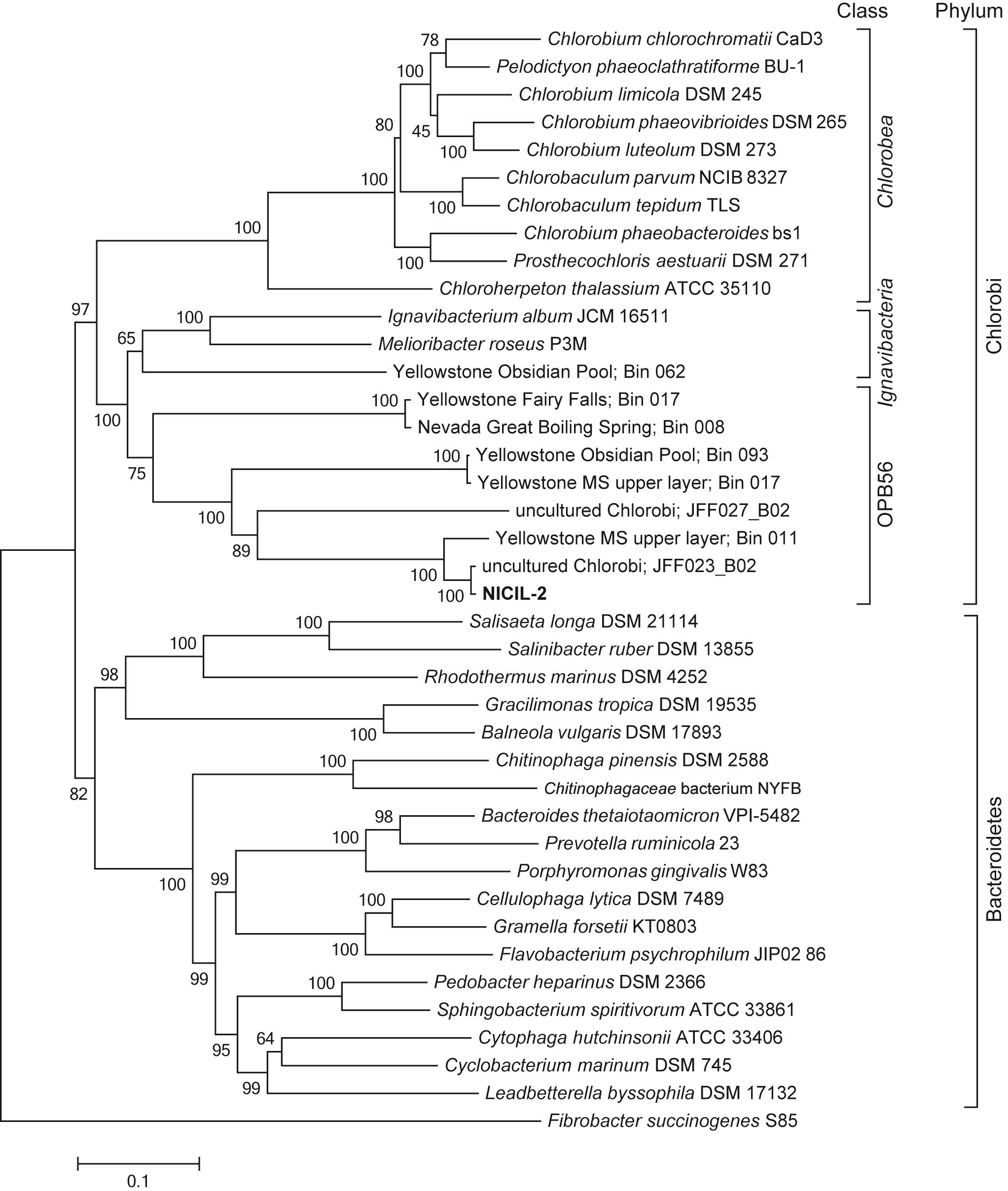
Microbial Communities Genomics Data
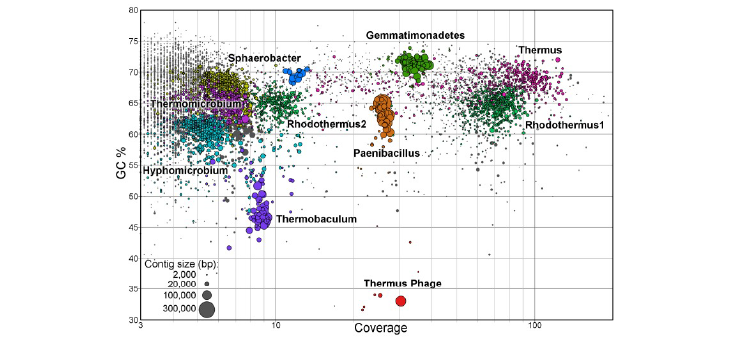
Adapting microbial communities to grow on biomass substrates has provided access to the genomes of uncultivated bacteria often overlooked in natural ecosystems. In this paper in ISME Journal, Hiras et al. (doi:10.1038/ismej.2015.158) adapted a microbial consortium to grow on ionic-liquid pretreated switchgrass as its sole carbon source. Among the most abundant bacteria is this community was a member of the OPB56 clade, an enigmatic, deep-branching lineage in the Chlorobi phylum. Reconstruction of its genome using MaxBin (click here) demonstrated that it grew on small organic molecules released by primary degrader in the microbial community. This reconstructed OPB56 genome was used to identify related genomes present in samples thermal springs in Yellowstone National Park and the Great Basin in Nevada, available here.
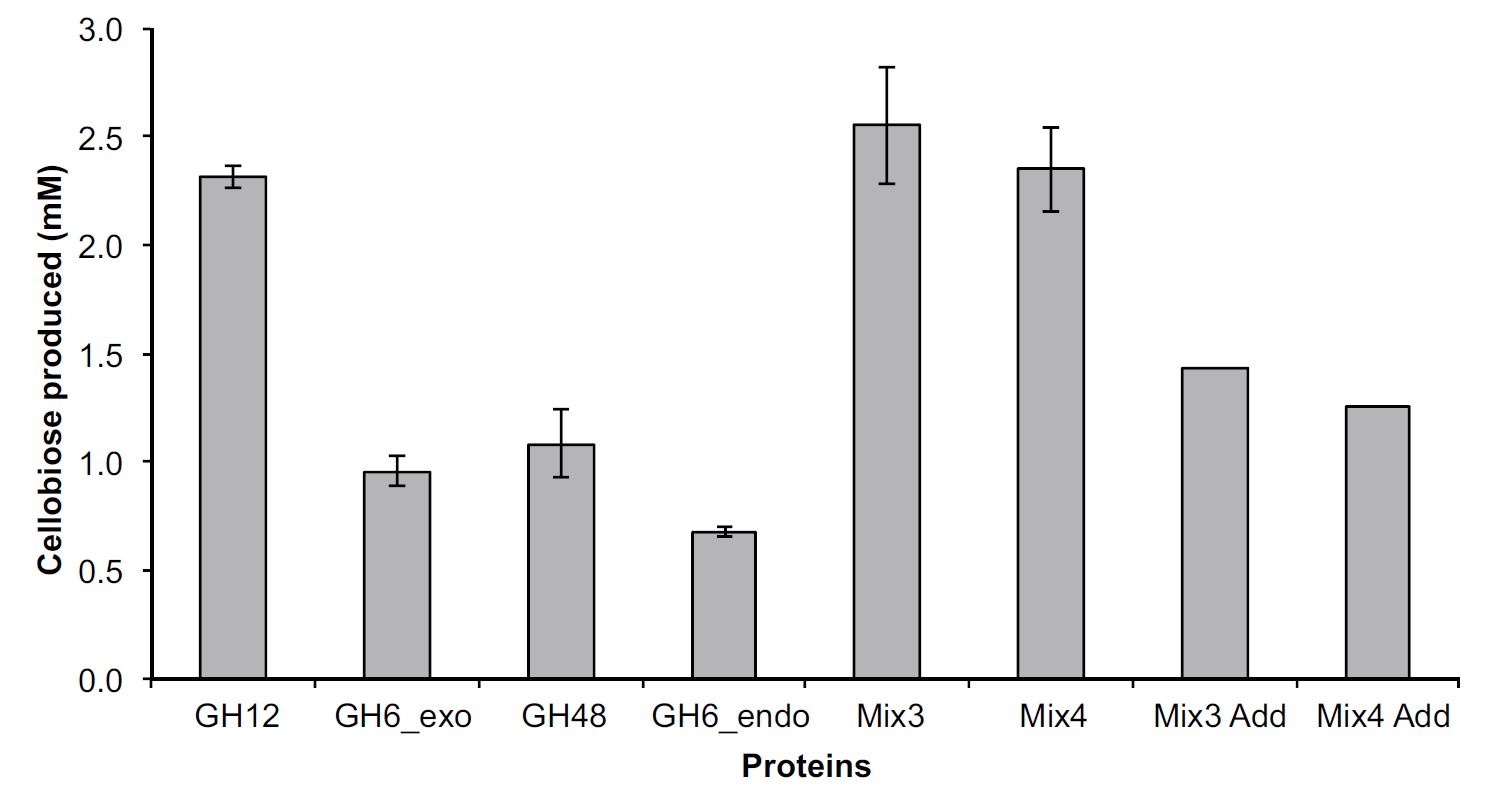
Comparative Community Proteomics
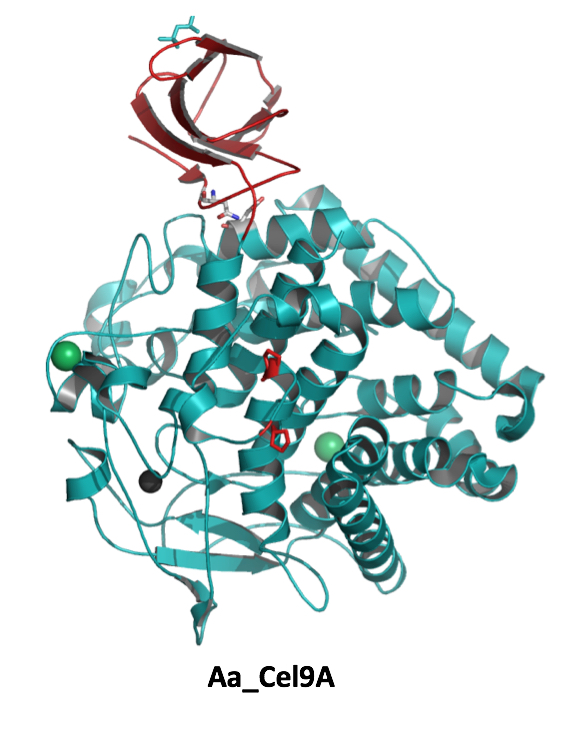
Comparative community proteomics has been used to identify a new role for a cellulase in the hydrolysis of crystalline cellulose. In this paper in mBio, Hiras et al. (doi: 10.1128/mBio.01106-16), the cellulase activities of two microbial consortia adapted to grow on crystalline cellulose diverged. A combination of genomic (available here, here, here and here) and proteomic analyses indicated a glycoside hydrolase family 12 protein was critical in the high activity of one consortium. These observations were supported by heterologous expression and biochemical characterization of the abundant cellulases produced by the community.
Relationship of thermo-tolerance to [C2C1Im][OAc]-tolerance
In Data et al., (Datta et al.), we compared the activity of cellulases from thermophilic organism to the activity of a cellulase from a mesophilic organism and showed the enzymes from the hypertheromophiles maintained higher levels of activity relative to their activity in the absence of [C2C1Im][OAc]. In Gladden et al., we characterized twenty-one cellulose-degrading enzymes derived from a thermophilic microbial community and found that 70% of them were stable and active in [C2C1Im][OAc]. A comparison of optimum temperature and activity in increasing concentrations of [C2C1Im][OAc] demonstrates that cellulases with higher optimum temperature are more likely to remain active in higher concentrations of [C2C1Im][OAc], further strengthening the link between thermotolerance and [C2C1Im][OAc]-tolerance for glycoside hydrolases.
Relationship of halo-tolerance to IL-tolerance
Due to their adaptation to high salinity environments, in the paper by Zhang et al., we hypothesized that halophiles may be a good source of IL-tolerant cellulases. Using a genome-based approach, we identified and characterized a halophilic cellulase (Hu-CBH1) from the halophilic archaeon, Halorhabdus utahensis. Hu-CBH1 is a heat tolerant haloalkaliphilic cellulase and is active in salt concentrations up to 5 M NaCl and is tolerant to high levels (20percent w/w) of several ILs, including [C2C1Im][OAc].
Phylogenetic guided discovery of [C2C1Im][OAc]-tolerant GH1 family enzymes
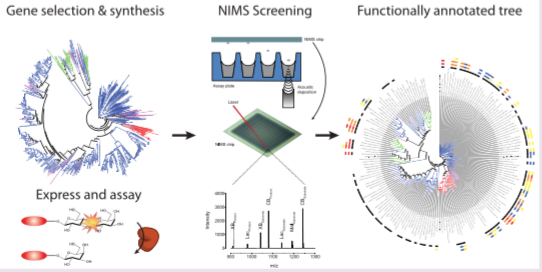
In the work of Heins et al., we synthesized and expressed 175 GH1s, selected from over 2000 candidate sequences to cover maximum sequence diversity, and assayed them over a range of temperatures and pHs using nanostructure- initiator mass spectrometry (NIMS, Deng et al.). We then identified GH1 enzymes active at 70 °C and 20% (v/v) [C2C1Im][OAc] over the course of a 24-h saccharification reaction. Using our unbiased approach, we identified several enzymes of different phylogentic origin having these desired properties.