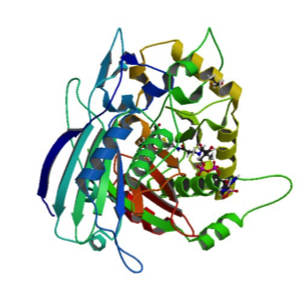
Structural Biology PvHCT Diffraction Data
The crystal structure of the enzyme PvHCT from switchgrass (Panicum vulgare) in complex with coenzyme A (CoA) and p-coumaroyl shikimate (5FAL) gives insight into its catalyzing action during the early steps of lignin biosynthesis. Moreover, the structure of PvHCT in complex with p-coumaroyl-CoA and protocatechuate (5FAN) highlights the substrate promiscuity of the enzyme towards non-canonical acceptors. The lignin pathway has received great attention because its engineering in bioenergy crops offers the potential to reduce the cost of biofuels and bioproducts. Raw diffraction datasets for this PvHCT structure are available for both 5FAL and 5FAN. Eudes A, et al., 2016, contains more information on the activity and flexibility of this enzyme.
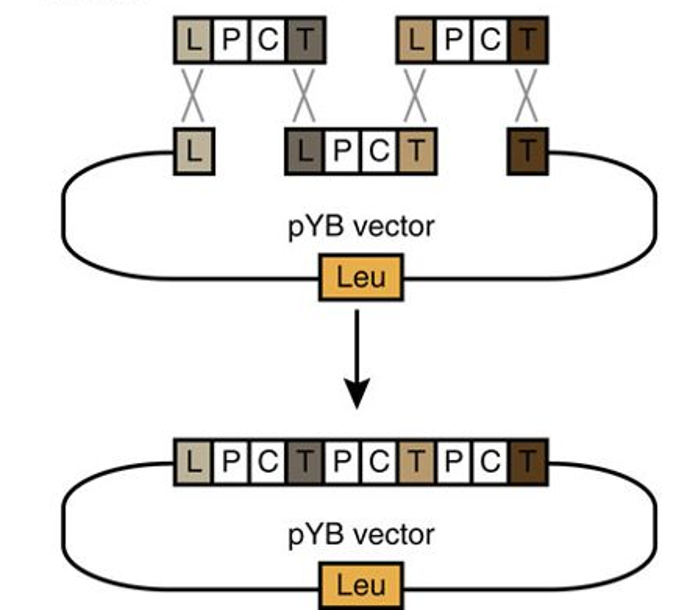
J-Stack v1.0 parts list
J-Stack is a method for rapidly assembling large plant synthetic biology constructs using yeast. The paper describing the methods can be found here: Shih et al. 2016. The synthetic parts (promoters, terminators, linkers etc.) are all available for distribution via the JBEI registry. The list of the parts, along with the JBEI registry identifiers, is found here: JStackv1 Parts List. Contact JBEI’s Plant Biosystems Design group for more information.
Structural Biology OsSERK2 Diffraction Data
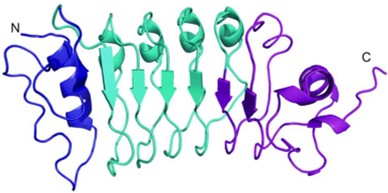
The rice cell wall receptor kinase SERK2 (OsSERK2) is critical for grass immunity, growth and development. The crystal structure (data is available here and here) provides insights into the mechanism of OsSERK2 function. Further information can be found in McAndrew R, et al., 2014.
Switchgrass Proteomics Data
The North American native prairie grass Panicum virgatum L. (switchgrass) is a promising bioenergy feedstock. The Joint Genome Institute is currently sequencing the Alamo AP13 cultivar, which has an estimated to have an effective genome size of 1600 Mbp. We have carried out proteomic analysis of this cultivar as a resource for the community to help develop switchgrass biology. Initial work on coleoptile endomembranes was published here and the data is deposited here. We have since extended this to soluble and membrane proteins for Alamo roots, leaves, stems and flowers in collaboration with Josh Heazlewood at the University of Melbourne and Ben Schwessinger at ANU. These data are currently unpublished, but available to researchers under the data usage principles established under the Ft Lauderdale/Amsterdam agreement. Please contact JBEI’s Plant Systems Biology group for access to the data.
Systems Biology Genome Sequences
Next generation sequencing is being used to discover the whole-genome sequence variation in 1001 strains of Arabidopsis thaliana. Six strains were assembled from pair-end Illumina sequence reads against TAIR10 by the Joint Genome Institute. The data for two more strains (Bay-0 and Sha) were recently realigned against the Arabidopsis Col-0 genome release TAIR10.
Systems Biology FT-NIR Spectra
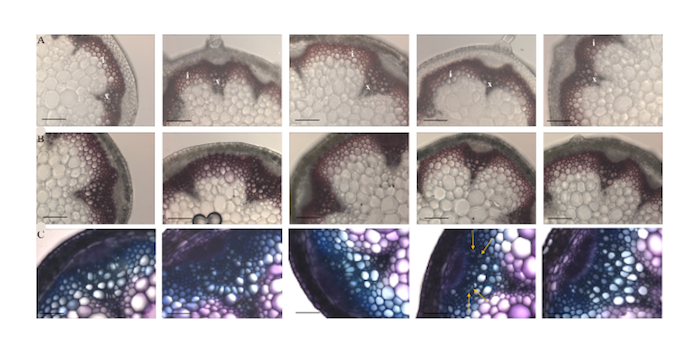
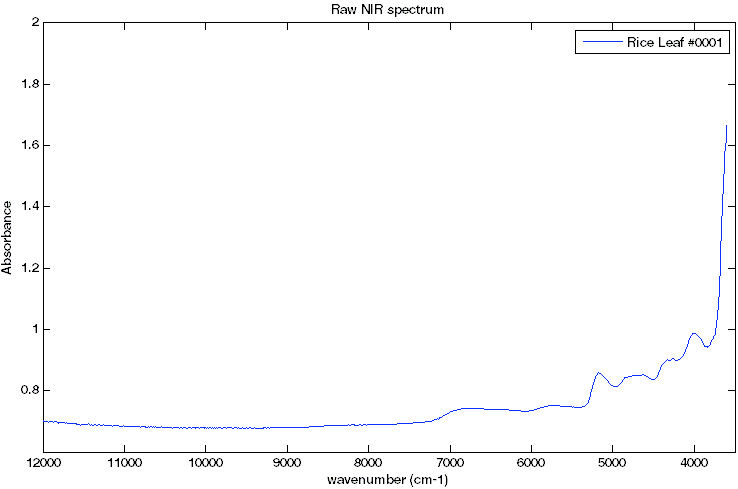
A rice mutant population was generated by fast neutron bombardment of the rice line Kitaake-Ubi-Xa21. Rice leaves were harvested at 3 weeks and analyzed using Fourier Transform Near Infrared Spectroscopy. These spectra were used in a pilot study to model cell wall sugar composition (Smith-Moritz AM, et al., 2011).
Systems Biology Golgi Proteomic Dataset
Biosynthesis of cell wall matrix polysaccharides takes place in the Golgi. Intact Golgi membranes were purified from Arabidopsis using density centrifugation and free-flow electrophoresis (FFE). Liquid chromatography-tandem mass spectroscopy was used to identify Golgi proteins. Identifications were made with one or more significantly matching peptides by Mascot (p < 0.05) against the Arabidopsis protein database TAIR10 (Parsons HT, et al., 2012).
Systems Biology Cytosolic Proteomic Dataset
The cytosolic cell fraction from Arabidopsis thaliana was isolated and analyzed using offline strong cation exchange liquid chromatography and liquid chromatography-tandem mass spectroscopy. Over one thousand proteins were identified with one or more significantly matching peptides against TAIR9 (Ito J, et al., 2011).
Cell Wall Engineering Microarray Analysis
In order to improve industrial processing, lignin monomers were over-expressed in Arabidopsis. Biosynthesis of these monomers is accomplished by expressing a bacterial enzyme in the plant stems. The resulting strain (IRX5:HCHL) was grown and assayed (Eudes A, et al., 2012). Microarray analysis was performed on complete Arabidopsis thaliana transcriptome microarrays containing 24,576 gene-specific tags (GSTs) corresponding to 22,089 genes from Arabidopsis.