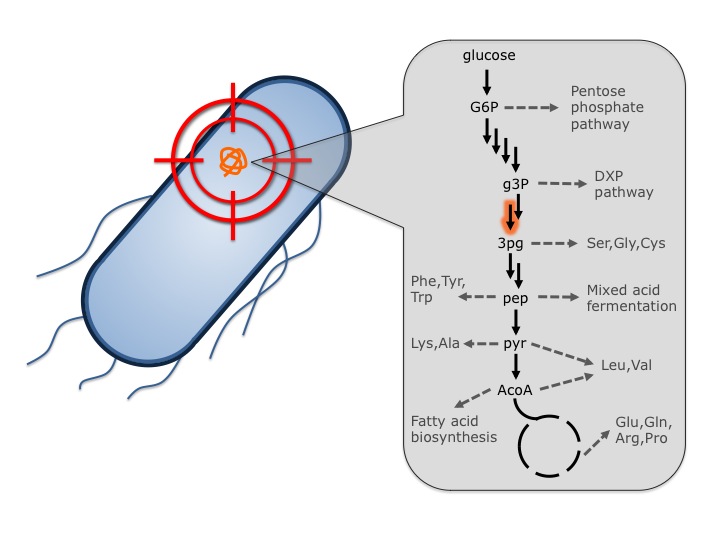
Access structural information, both raw diffraction data and PDB entries, for proteins that could be optimized to improve the production of advanced biofuels and bioproducts.
(Please note: The 4EWP, 3MMW, and 3EZ8 raw diffraction datasets consist of 180 images approximately 8 MB in size; 3 MMU has 270 images of the same size. 3U48, 3U4A, 3SAE, 3SDR, 3SDT, 3SDU, and 3SDV datasets contain approximately 360 images that are 19 MB; 3SDQ has 720 images, 19 MB each.)
Structural Biology – Crystal structure of EilR transcription factor
Tightly regulated promoters are essential for numerous biological applications, where strong inducibility, portability, and scalability are desirable. Structural biology group provides molecular insights into specific interactions of EilR with its operator (PDB ID 5VL9) and with two inducers, crystal violet (PDB ID 5VLM) and malachite green (PDB ID 5VLG). The versatility of an expression system named Jungle Express opens the way for tightly controlled and efficient gene expression that is not restricted to host organism, substrate, or scale. Ruegg et al., 2018, contains more information on EilR structural study.
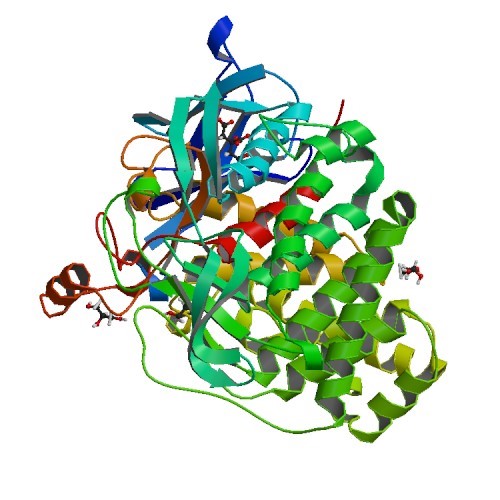
Structural Biology – Crystal structure of GH family 9 endoglucanase J30
The development of robust enzymes, in particular cellulases, is a key step in the success of biological routes to `second-generation’ biofuels. The crystal structure of wild-type J30 (PDB ID 5U0H) is presented along with that of a designed triple-mutant variant (PDB ID 5U2O) with improved characteristics for industrial applications. Through the introduction of a structural Zn2+ site, the thermal tolerance was increased by more than 10°C and was paralleled by an increase in the catalytic optimum temperature by more than 5°C. Ellinghaus et al., 2018, for further details.
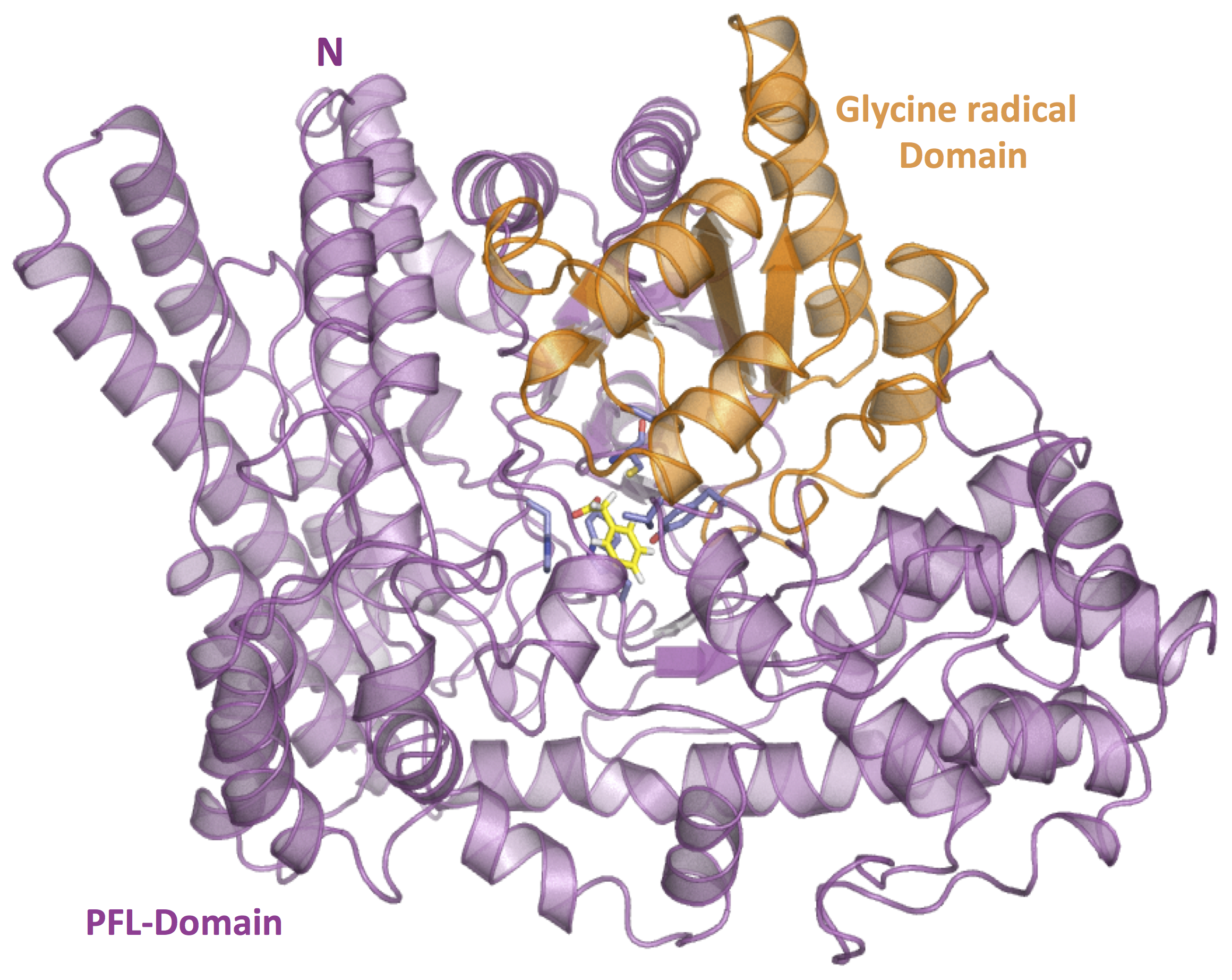
Structural Biology – Molecular model of PhdB in complex with Phenylacetate
Microbial toluene biosynthesis was reported in anoxic lake sediments more than three decades ago, but the enzyme catalyzing this biochemically challenging reaction has never been identified. The structural biology group built the toluene-producing enzyme PhdB molecular model in complex with phenylacetate substrate Further information can be found in Beller et al., 2018.
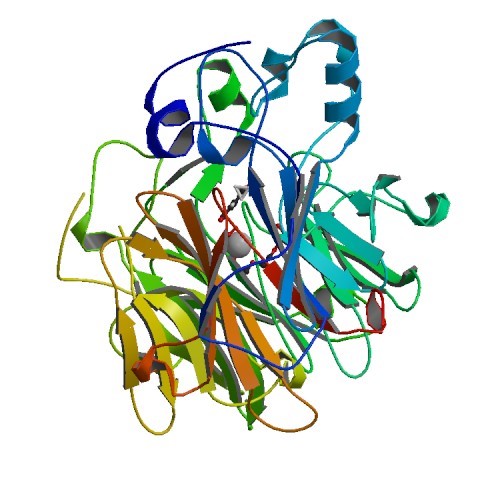
Structural Biology – Crystal structure of NOV1, a Resveratrol-Cleaving Dioxygenase
Stilbenes are diphenyl ethene compounds produced naturally in a wide variety of plant species and some bacteria. Stilbenes are also derived from lignin during kraft pulping. Stilbene cleavage oxygenases (SCOs) cleave the central double bond of stilbenes, forming two phenolic aldehydes. The structural biology group solved the structures of an SCO called NOV1 in an apo-form (PDB ID 5J53), in complex with its substrate resveratrol (PDB ID 5J54) and its product vanillin (PDB ID 5J55). NOV1 cleaves a wide range of stilbene-like compounds with a 4′-OH group, offering potential in processing some solubilized fragments of lignin into monomer aromatic compounds. Further information can be found in McAndrew et al., 2016.
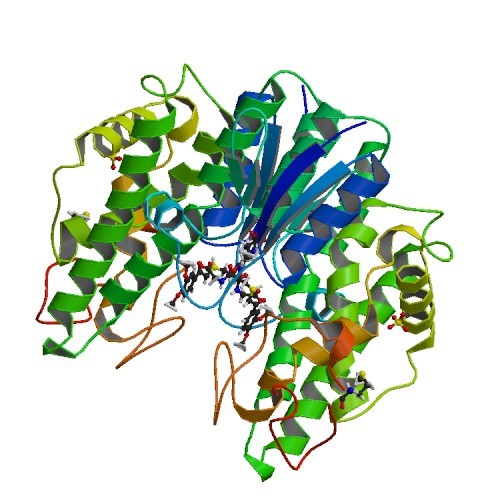
Structural Biology – Structural Characterization of the Lignin β-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6
Biological cleavage of lignin has been well characterized in fungi, in which enzymes that create free radical intermediates are used to degrade this material. In contrast, a catabolic pathway for the stereospecific cleavage of β-aryl ether units that are found in lignin has been identified in Sphingobium sp. SYK-6 bacteria. The crystal structures and biochemical characterization of the NAD-dependent dehydrogenases [LigD (PDB IDs 4Y98 4Y9D), LigO (PDB IDs 4YA6 4YAC), and LigL (PDB IDs 4YAE 4YAG 4YAI)] and the glutathione-dependent lyase LigG (PDB IDs 4YAP 4YAV) provide new insights into the early and late enzymes in the β-ether degradation pathway. Refer to Pereira et al., 2016, for further details.
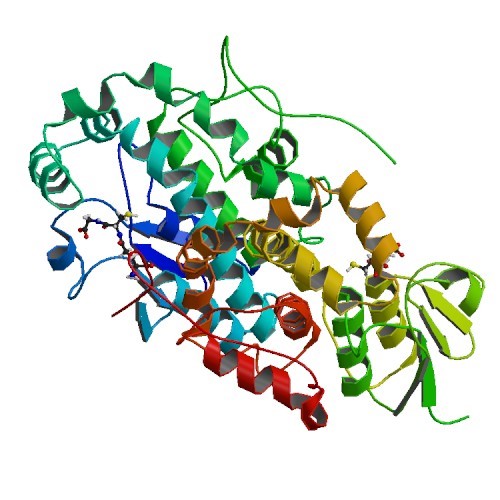
Structural Biology – Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of β-Aryl Ether Bonds in Lignin
Lignin is a combinatorial polymer comprising monoaromatic units that are linked via covalent bonds. Although lignin is a potential source of valuable aromatic chemicals, its recalcitrance to chemical or biological digestion presents major obstacles to both the production of second-generation biofuels and the generation of valuable coproducts from lignin’s monoaromatic units. The structural biology group of JBEI in collaboration with Great Lakes Bioenergy solved the x-ray crystal structures of the glutathione-dependent β-etherases, LigE (PDB IDs 4YAM 4YAN) and LigF (PDB ID 4XT0). Refer to Helmich et al., 2016, for further details.
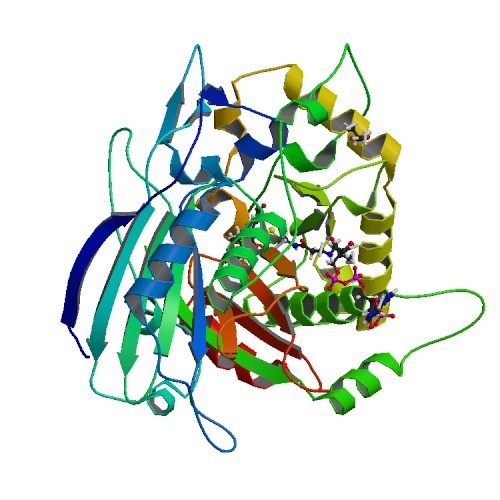
Structural Biology – Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin
Lignin poses a major challenge in the processing of plant biomass for agro-industrial applications. Hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase (HCT) is a key metabolic entry point for the synthesis of the most important lignin monomers: coniferyl and sinapyl alcohols.The structural study of HCT called PvHCT2 confirmed the binding of a non-canonical acceptor in a similar manner to shikimate in the active site of the enzyme (PDB IDs 5FAL 5FAN). Refer to Eudes et al., 2016, for further details.
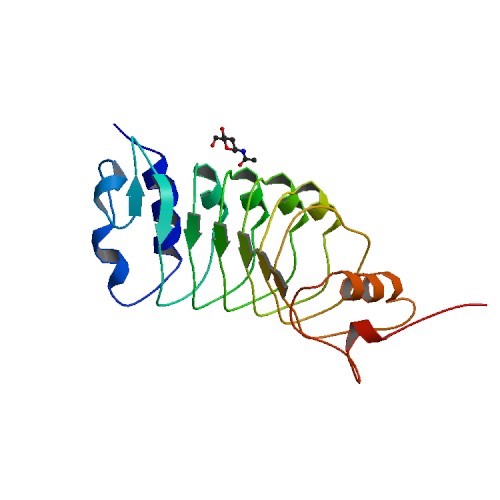
Structural Biology – Structure of the OsSERK2 leucine-rich repeat extracellular domain
Somatic embryogenesis receptor kinases (SERKs) are leucine-rich repeat (LRR)-containing integral membrane receptors that are involved in the regulation of development and immune responses in plants. The crystal structures of the LRR domains of OsSERK2 (PDB ID 4Q3G) and a D128N OsSERK2 mutant (PDB ID 4Q3I) were solved. These structures suggest that the aspartate mutation does not generate any significant conformational change in the protein, but instead leads to an altered interaction with partner receptors. Further information can be found in McAndrew et al., 2014.
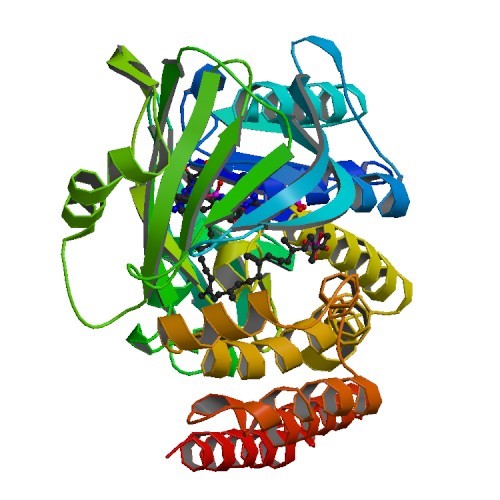
Structural Biology – Constructing tailored isoprenoid products by structure-guided modification of geranylgeranyl reductase
The archaeal enzyme geranylgeranyl reductase (GGR) catalyzes hydrogenation of carbon-carbon double bonds to produce the saturated alkyl chains of the organism’s unusual isoprenoid-derived cell membrane. The structural biology group solved the crystal structures of apo-form GGR (PDB IDs 4OPL) from Sulfolobus acidocaldarius, including the structure of GGR bound to geranylgeranyl pyrophosphate (GGPP) (PDB IDs 4OPT 4OPU). Further information can be found in McAndrew et al., 2014.
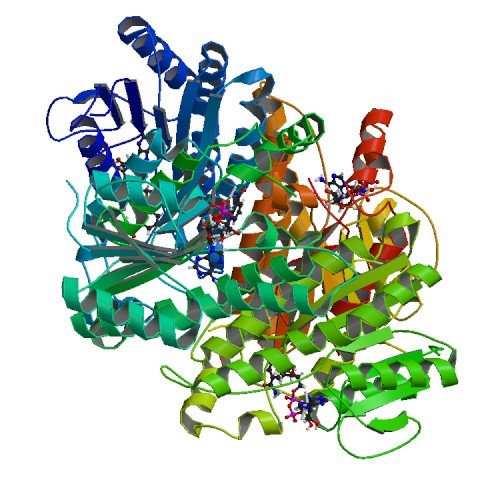
Structural Biology – Biochemical and structural studies of NADH-dependent FabG used to increase the bacterial production of fatty acids under anaerobic conditions
Major efforts in bioenergy research have focused on producing fuels that can directly replace petroleum-derived gasoline and diesel fuel through metabolic engineering of microbial fatty acid biosynthetic pathways. Typically, growth and pathway induction are conducted under aerobic conditions, but for operational efficiency in an industrial context, anaerobic culture conditions would be preferred to obviate the need to maintain specific dissolved oxygen concentrations and to maximize the proportion of reducing equivalents directed to biofuel biosynthesis rather than ATP production. A major concern with fermentative growth conditions is elevated NADH levels, which can adversely affect cell physiology. The purpose of this project was to identify homologs of Escherichia coli FabG, an essential reductase involved in fatty acid biosynthesis, that display a higher preference for NADH than for NADPH as a cofactor. Four potential NADH-dependent FabG variants were identified through bioinformatic analyses supported by crystal structures solved between 1.3- to 2.0-Å resolution (PDB IDs 4NBW 4NBV 4NBU 4NBT). Refer to Javidpour et al, 2014, for further details.
Structural Biology – FabH Diffraction Data
The crystal structure of the enzyme FabH (4EWP) from the bacterium, Micrococcus luteus, provides evidence for its catalyzing action during the first step of fatty acid biosynthesis. This pathway has gained attention because fatty acid molecules are favorable precursors for compounds that could substitute for petroleum-derived fuels. Raw diffraction images for this FabH structure can be accessed here. Pereira JH, et al., 2012, contains more information on the activity and selectivity of this molecule.
Structural Biology – Cel5A Diffraction Data
Thermatoga maritima Cel5A is a thermally stable glycoside hydrolase and is an excellent candidate for use in the degradation of polysaccharides present on biomass. The Tm_Cel5A structural information (PDB IDs 3MMU and 3MMW) can be used to potentially increase the thermostability of mesophilic cellulase enzymes. Raw diffraction datasets are available for both 3MMU and 3MMW. Refer to Pereira JH, et al., 2010, for further details.
Structural Biology – Cel9A Diffraction Data
Endoglucanase Cel9A from the heat- and acid-loving bacteria, Alicyclobacillus acidocaldarius, is used to break down ?-1,4-linked glucans in cellulose. The crystal structure (3EZ8) provides insight into the catalytic mechanism, which could help to optimize process of degrading biomass and the producing of sugars for subsequent fermentation to fuel. Raw diffraction datasets are available for download. Further information can be found in Pereira JH, et al., 2009.
Structural Biology – Glucosidase Diffraction Data
A microbial enzyme that could be useful in the pretreatment process of second-generation biofuels was identified from metagenomic analysis of a switchgrass-adapted compost community. The crystal structure of this 3-?-glucosidase was solved in complex with glucose (3U48) and a variant was crystallized with cellopentaose (3U4A). See McAndrew RP, et al., 2013, for a description of this enzyme’s novel structure. Download the raw diffraction datasets here: 3U48, 3U4A.
Structural Biology – AgBIS Diffraction Data
The sesquiterpene bisabolene is a precursor to bisabolane, an advanced biofuel. Abies grandis alpha-bisabolene (AgBIS) was used to produce high-titer microbial bisabolene. This sesquiterpene synthase was crystallized in the apo form (3SDQ) and with 5 inhibitors (3SAE, 3SDR, 3SDT, 3SDU, 3SDV). Learn more about this enzyme structure in McAndrew RP, et al., 2011. or by accessing the raw diffraction images (3 SDQ, 3SAE, 3SDR, 3SDT, 3SDU, 3SDV).
JBEI Targeted Proteomic Methods and Data
Targeted proteomic methods and data for JBEI articles can be found on the Panorama Repository Software for Targeted Proteomics Assays from Skyline.
JBEI Technology Mass Spectrometry Datasets
Mass Spectrometry Imaging (MSI) data for JBEI Technology publications can be found on OpenMSI, a web-based platform for the visualization of Mass Spectrometry Imaging data.