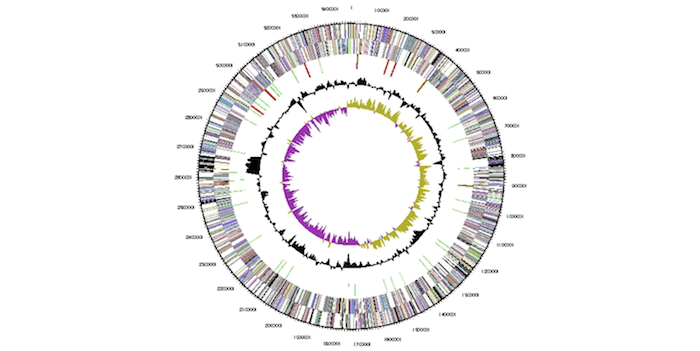
Multiple annotated DNA sequences, DNA design and assembly files, primers, plasmids and strains have been produced, validated, and shared. Explore these and microarray datasets for yeast responses to biofuel-related stress. Image from Chertkov O, et al., Stand. Genomic Sci. 5:1 (2011).
Functional genomics for strain engineering
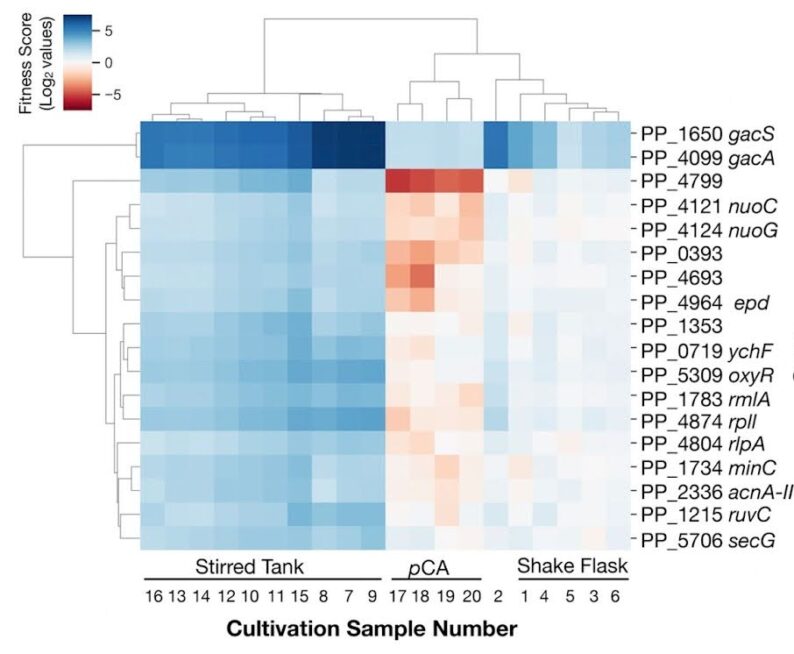
Developing improved microbial strains for bioconversion routinely uses functional genomics data from RNA-seq, Proteomics and RB-TnSeq.Eng et. al., 2021 evaluated P. putida KT2440 in a bioreactor using RB-TnSeq to identify genes that provide fitness. The RB-TnSeq P. putida reactor data is available from the fitness browser. Banerjee et al 2020 showed the use of RNA-seq data (from the Join Genome institute) to curate Genome Scale Metabolic Models. All RNA-seq data from this study is available via NCBI Bioproject PRJNA580539 and also the JGI Gold site.
Genome sequence of bioproduction strains
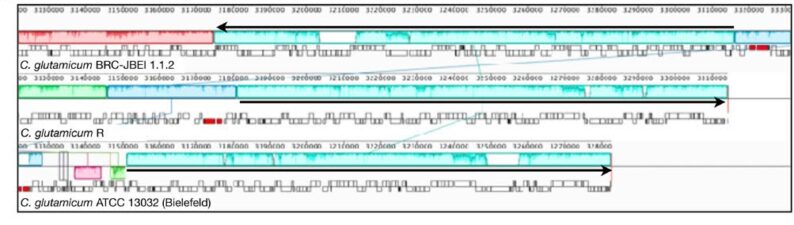
Research at JBEI uncovered a strain variant of the commonly used Corynebacterium glutamicum strain; C. glutamicum BRC-JBEI 1.1.2 that was used to engineer the production of isoprenol (Sasaki et. al., 2019), TMP (Srinivasan et. al., 2024) and most recently to develop robust additional tools for bioproduction (Luckie et. al., 2024). The BRC-JBEI 1.1.2 was resequenced at the JGI and is available via NCBI Genome Assembly ID: GCA_011761195.1. Additional omics data for BRC-JBEI 1.1.2 tolerance to Cholinium lysinate is also available from NCBI listed in Banerjee et al 2021.
JBEI ALE data
 To develop improved microbial chassis for bioproduction, JBEI researchers use ALE to obtain strain with improved tolerance to deconstruction reagents (), catabolism of aromatics (), sugars (). When high throughput automated ALE is used, routine sequencing allows us to capture genome wide mutations that accumulate in evolved strain lines. These data are organized and available via the ALE DB. Here are links to recent projects that led to publications:
To develop improved microbial chassis for bioproduction, JBEI researchers use ALE to obtain strain with improved tolerance to deconstruction reagents (), catabolism of aromatics (), sugars (). When high throughput automated ALE is used, routine sequencing allows us to capture genome wide mutations that accumulate in evolved strain lines. These data are organized and available via the ALE DB. Here are links to recent projects that led to publications:
- Pputida_carbon_ALE – 10.1021/acssuschemeng.1c03765
- Pputida_carbon_ALE_2 – 10.1021/acssuschemeng.1c03765
- P. putida Hydroxycinnamic TALE – 10.1016/j.mec.2020.e00143
- ILTolerance – 10.1186/s12934-017-0819-1
High-throughput and automated DBTL cycle for polyketide synthase reprogramming: Retrobiosynthesis of unnatural lactams
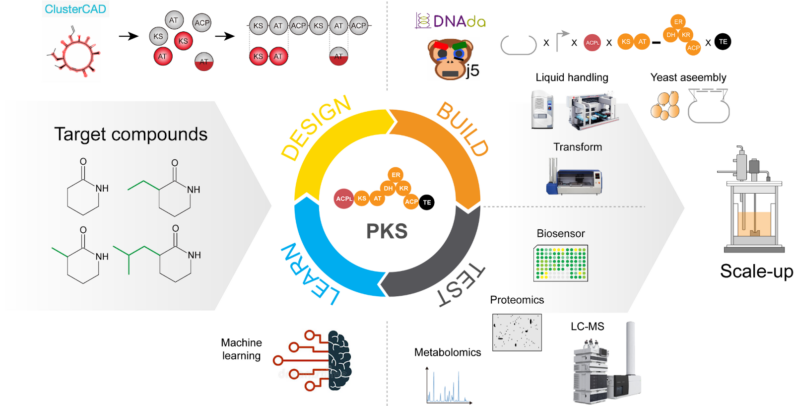
High-throughput and automated DBTL cycles were developed for PKS-based retrobiosynthesis, as partially demonstrated by Nava et al., 2023. Unnatural lactams were selected as the initial target compounds and successfully produced from biomass using P. putida KT2440. Hundreds of chimeric PKSs were constructed by exchanging domains or modules at various junction sites to produce unnatural lactams. The resulting production data will be presented in an upcoming paper.
Biosynthetic platforms for industrial diols and derivatives
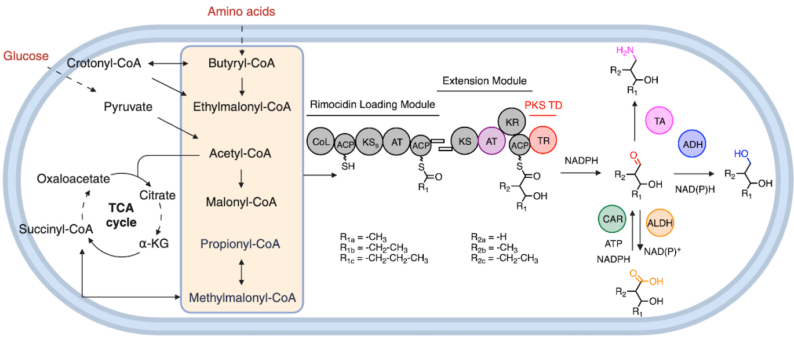
Developing retrobiosynthetic platforms suitable for biomanufacturing petroleum-derived industrial molecules. Using integrative approaches in genome mining, biochemistry, structural biology and metabolic engineering, Dan et al. identified hundreds of candidate polyketide synthase (PKS) thioreductases (TRs), characterized PKS TR catalysis, incorporated TRs into assembly-line PKS engineering, and enabled biosynthetic access to seventeen diols and derivatives in Streptomyces albus. These products include 1,3-butanediol (humectant) and 2-ethyl-1,3-hexanediol (insect repellent). The coordinates of CpkC TR crystal structure are available on Protein Data Bank (PDB 8V1X). All proteomics data from this study are also available via the ProteomeXchange Consortium (PXD055149).
Functional genomics for strain engineering
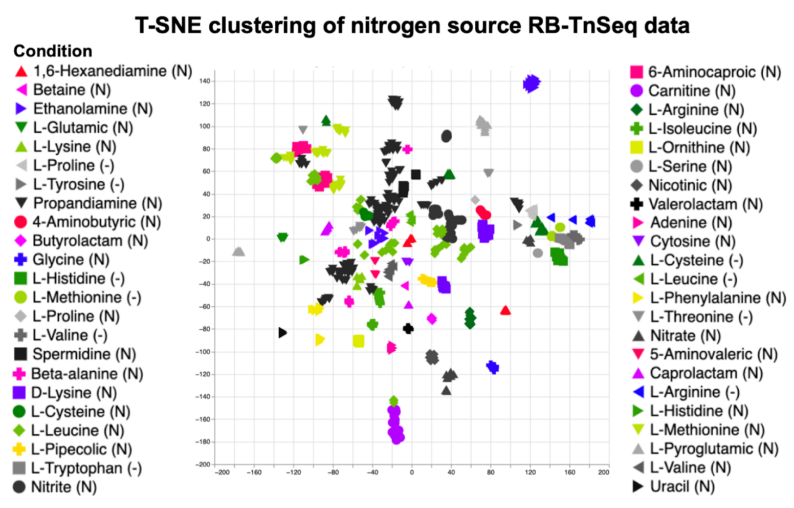
To better understand the metabolism of a JBEI host organism and aid in metabolic engineering efforts, JBEI researchers leveraged RB-TnSeq to obtain functional evidence for the genetic basis of nitrogen metabolism in Pseudomonas putida KT2440 (Schmidt and Pearson et al 2022). Researchers tested 52 nitrogen sources and 19 amino acid drop-out conditions, resulting in significant phenotypes in 672 different genes. The full dataset can be accessed through the fitness browser.
Improving the production of value-added products in different host organisms using codon optimization
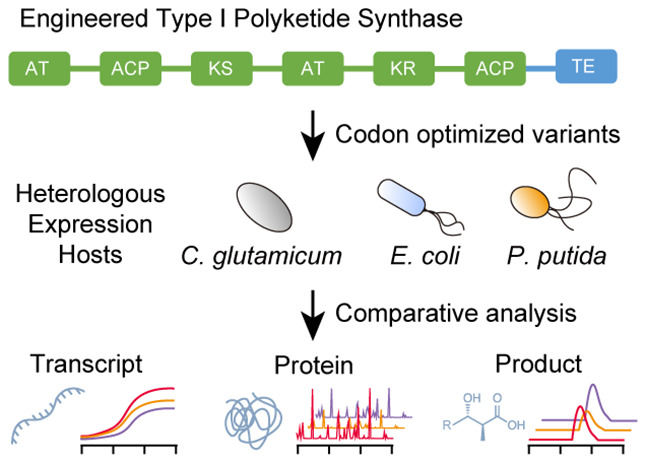
Protein engineering and heterologous expression in host organisms holds enormous potential as a rational production platform for the biosynthesis of specialty chemicals. However, choosing the wrong codon optimization strategy for expression in a heterologous host can have detrimental effects on protein and product levels. (Schmidt et. al., 2023) investigated the relationship between codon optimization strategies and the resulting transcript, protein, and product expression levels. By employing the strategies developed in their study, JBEI researchers successfully produced unnatural polyketides across three different hosts. This research also led to the development of a free, transparent, and customizable codon optimization tool, available at basebuddy.lbl.gov.
A bio-based platform for renewable and recyclable plastics with customizable properties
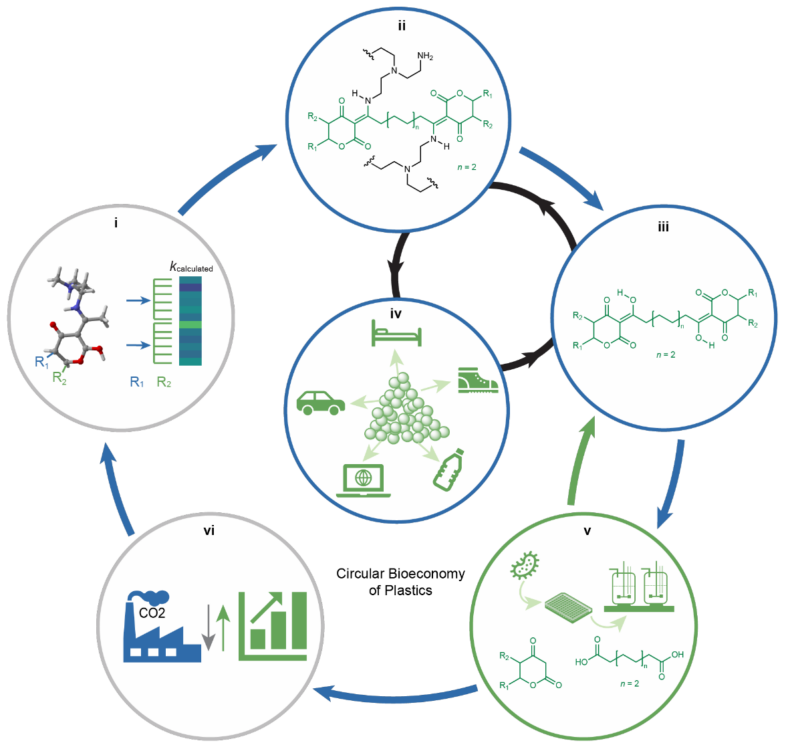
Traditional plastics are almost exclusively produced from non-renewable feedstocks and face considerable challenges in recycling. In this recent work by (Wang et. al., 2024), researchers took an interdisciplinary approach that combined computational materials design, polymer chemistry, synthetic biology, and systems analysis. This integrated effort enabled the design of customizable, biorenewable plastics with tailored properties, contributing to a circular bioeconomy. The microbial strains used to produce the monomers in this work are accessible through the JBEI Public Registry.
Adaptive Evolution of E. coli strains for Malonyl-CoA Production
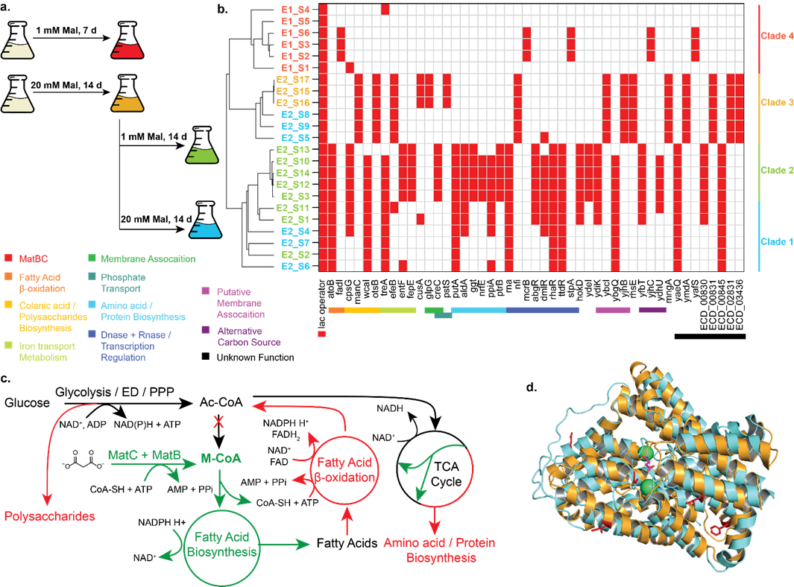
Heterologous expression of polyketide synthase (PKS) genes in Escherichia coli enables the production of valuable natural and synthetic products, but limited malonyl-CoA (M-CoA) availability hinders high-titer production. Introducing an orthogonal malonate transport and CoA ligation pathway significantly enhanced M-CoA-derived polyketide titers (Klass et al., 2024 ; Wang et al., 2023). Disrupting the native M-CoA pathway and reconstituting it through malonate supplementation enabled adaptive laboratory evolution (ALE), revealing mutations though whole genome sequencing and proteomic analysis that increased M-CoA and polyketide levels. This approach not only improves E. coli as a polyketide host but also provides insights into M-CoA modulation dynamics.
Archived Data:
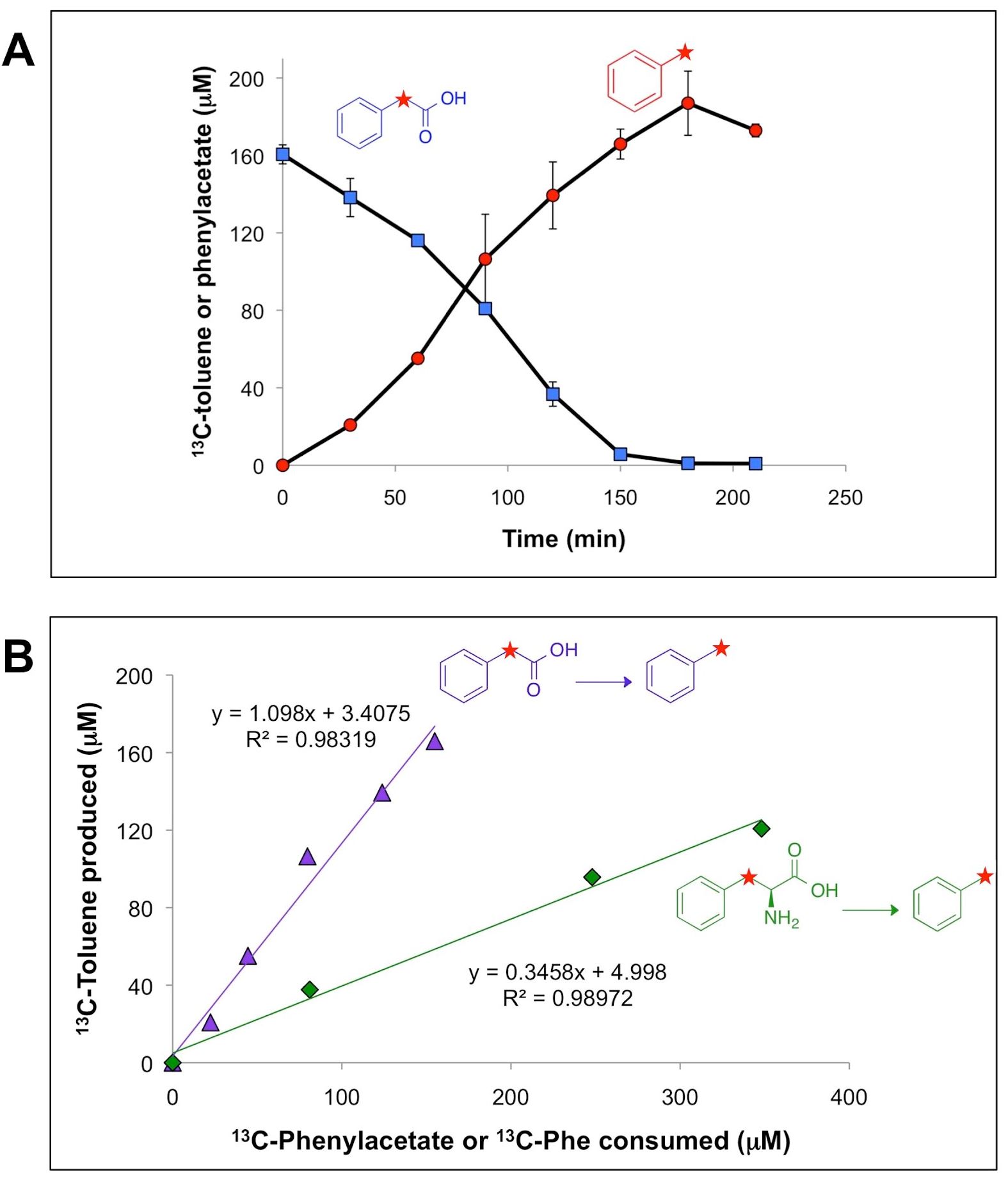
Biofuels Pathways Proteomic and 16s iTag data
Researchers at JBEI have investigated an enzyme that could enable first-time biochemical production of the widely used octane booster, toluene (Zargar et al. 2016). For this work, in vivo, in vitro, metagenomic, and metaproteomic studies were conducted with an anaerobic microbial community derived from sewage sludge that quantitatively produced toluene from phenylacetic acid (via decarboxylation). Data included community composition by 16S rRNA gene iTag analysis and metaproteomic analysis of partially purified fractions of the community proteome that catalyzed toluene biosynthesis from phenylacetic acid; proteins were identified by mapping peptides to a community metagenome (Joint Genome Institute IMG Taxon ID 3300001784).
Host Engineering Microarray Data
The yeast Saccharomyces cerevisiae can be used to convert biomass to biofuels. In order to better understand yeast response due to stressors that could inhibit or limit this process, yeast was evaluated under various conditions. Microarray analysis was performed at different time points to determine yeast genome-wide response when exposed to salt, acid, hypoxic conditions, and hydrogen peroxide, as well as ethanol, isopentenol, and other biofuels candidates.
Synthetic Biology Combinatorial Plasmids
PR-PR is an open-source biology friendly programming language that has utility as a cross-platform lab automation system. PR-PR can be used with liquid-handling robots, microfluidics, and microscopy, as well as for protocol translation into human languages. In the course of further developing PR-PR, researchers wrote protocols for processes to manipulate DNA: combinatorial modified Golden Gate DNA assembly, Kunkel DNA mutagenesis and hierarchical Gibson DNA assembly (Linshiz, et al., 2014). Information for initial and resulting materials is available in the JBEI registry*, including DNA sequences, annotated sequence and sequencing trace validation files, and design files.
Synthetic Biology Bacterial Strains & Plasmids
ScanDrop is an all-in-one microfluidic system for detecting and reporting the presence of bacteria in drinking water (Golberg, et al., 2014). Target bacteria were captured using antibody-coated magnetic beads and then encapsulated in picoliter-sized droplets with fluorescently labeled antibodies for detection. Plasmids expressing either red or green fluorescent proteins were transformed into a strain of E. coli and used to demonstrate the feasibility of this system. Specific information about these materials can be found in the public instance of the JBEI Registry*.
Synthetic Biology Bacterial Strains & Plasmids
Ralstonia eutropha is a bacterium that can grow using hydrogen and carbon dioxide in an aerobic environment. When food sources are limited, R. eutropha produces the biopolymer, polyhydroxybutyrate (PHB). A plasmid-based toolbox was developed to optimize and diversify production of hydrocarbons in these bacteria (Bi, et al., 2013). Users can log on to the public instance of the JBEI Registry* to view all 60 bacterial strains and plasmids, which contain a variety of origins of replication, promoters, and ribosomal binding sites.
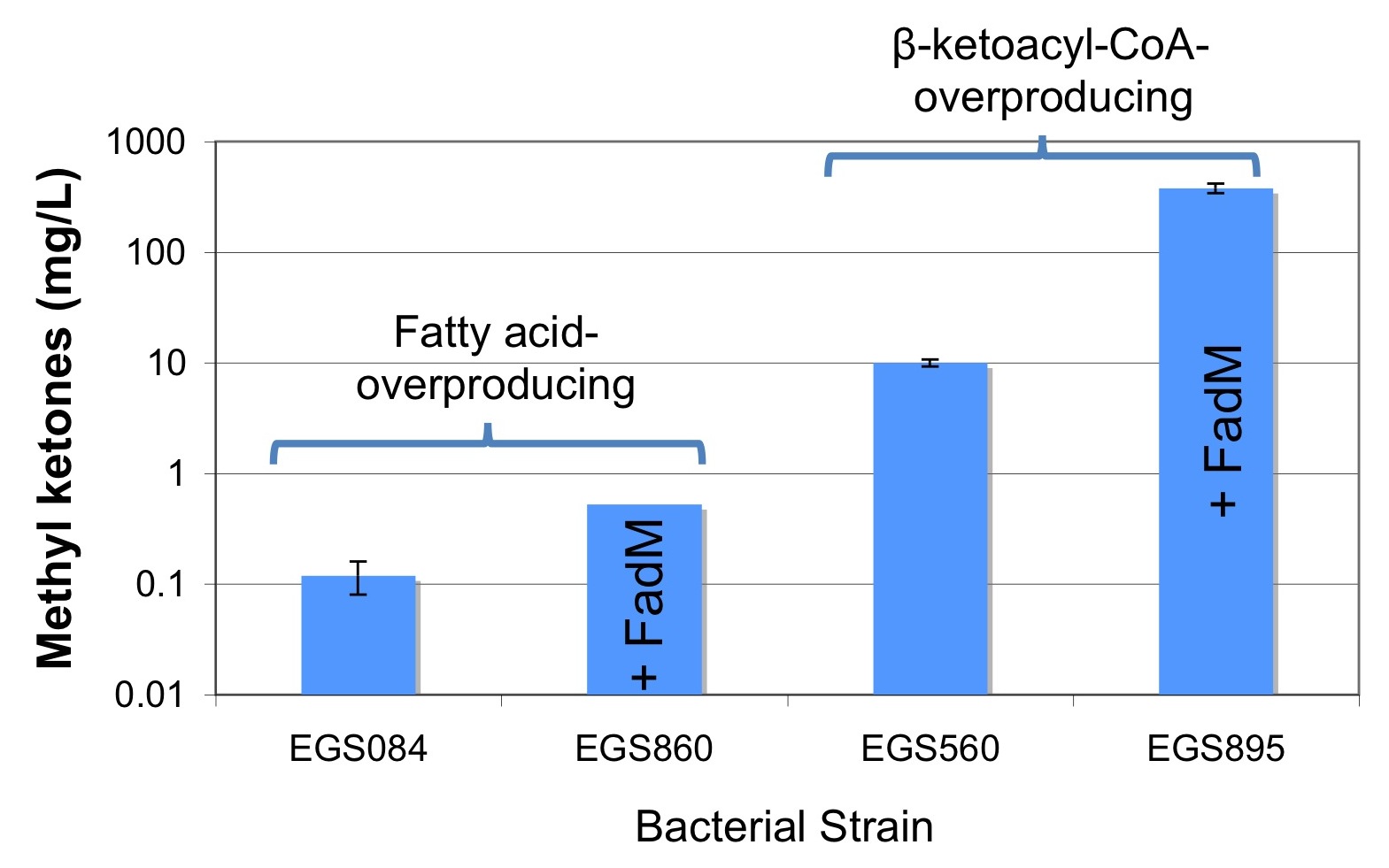
Biofuels Pathways Microarray Data
JBEI has engineered a new pathway for diesel-range methyl ketone biosynthesis in E. coli (Goh E-B, et al., 2012). Part of this effort involved discovery, through transcriptomic evidence (whole-genome microarrays), that up-regulation of the native fadM thioesterase in E. coli was correlated with enhanced methyl ketone titers.
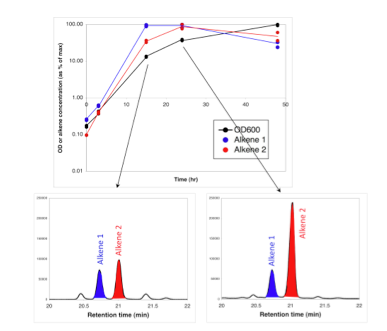
Biofuels Pathways Microarray Data
JBEI researchers identified three genes in the bacterium Micrococcus luteus that are associated with long-chain alkene biosynthesis from fatty acids (Beller H, et al., 2010). Related research (Pereira HJ, et al., 2012) used two different lines of evidence to explain the increase in the ratio of anteiso– to iso-branched alkenes that was observed during the transition from early to late stationary phase in M. luteus: structural studies of a key enzyme involved in fatty acid biosynthesis in M. luteus (FabH, or ß-ketoacyl-ACP synthase III) and transcriptional (whole-genome microarray) studies of M. luteus during different growth phases.
*Please note: Any user can create an account on the public instance of the JBEI Registry by going to https://public-registry.jbei.org/.