Monday, July 10, 2017
UC BERKELEY!
 On Monday, July 10th, instead of meeting at JBEI, Teacher Assistants Ron and Virginia met us at North Berkeley BART Station at around 9 AM. When we were all present, we began to walk down to UC Berkeley which was a 10 minute walk to get there and then maybe another 10 min walk to get to where we were supposed to be on campus. When we got there, we met about 10 undergraduates that were present to participate in our activity. Our activity was to build a rollercoaster with a few long pieces of styrofoam and two marbles. The goal was to receive the most points with loops, corkscrews and turn and to make the ball into the cup and stay there. This activity taught teamwork, collaboration, and some creativity. We then received a talk with a panel of engineers with different backgrounds which was interesting and inspiring. After that we walked around campus with Ron and Virginia showing us different things like the libraries. I took a picture with Mark Twain himself (Not Really). We had two other presentations, one from Kevin who gave us a background of UC Berkeley and another woman who gave us tips on getting into college. This experience brought to my eyes the life of a college student and the grind necessary to succeed and do what you want to do in your future.
On Monday, July 10th, instead of meeting at JBEI, Teacher Assistants Ron and Virginia met us at North Berkeley BART Station at around 9 AM. When we were all present, we began to walk down to UC Berkeley which was a 10 minute walk to get there and then maybe another 10 min walk to get to where we were supposed to be on campus. When we got there, we met about 10 undergraduates that were present to participate in our activity. Our activity was to build a rollercoaster with a few long pieces of styrofoam and two marbles. The goal was to receive the most points with loops, corkscrews and turn and to make the ball into the cup and stay there. This activity taught teamwork, collaboration, and some creativity. We then received a talk with a panel of engineers with different backgrounds which was interesting and inspiring. After that we walked around campus with Ron and Virginia showing us different things like the libraries. I took a picture with Mark Twain himself (Not Really). We had two other presentations, one from Kevin who gave us a background of UC Berkeley and another woman who gave us tips on getting into college. This experience brought to my eyes the life of a college student and the grind necessary to succeed and do what you want to do in your future.
Tuesday, July 11, 2017
My Thumb Hurts
Today we had one of our busiest days of the entire program (I wasn’t ready). We were given our bacteria that was put into an LB and CMC liquid media and put them into a flask. We all had three to do so we had nine bacterial eppendorf tubes that we had to prepare. We had one 1:1 dilution, 1:2 dilution, and a negative solution. The negative solution is a solution that is for sure not going to react which is used as a reference. The 1:1 and the 1:2 dilution were diluted with MES (2-ethanesulfonic acid) which we used as a buffer. We then put it into all three of the 2 mL eppendorfs. 9 eppendorfs in total (3 eppendorfs per bacterial sample). We then put the two dilutions into the centrifuge and put the negative into the 95o incubator to kill the bacteria and make it truly “negative”. After doing that for all nine of the eppendorfs ready, we then put the supernatant of the solutions into a 1.7 ml eppendorf and mixed with CMC as another buffer. When mixing all of the solutions along with the negatives, we then put them into the PCR plates following the chart that was made for us by Raphael (El), one of the post-docs who helped us. After pipetting all of our solutions along with the standards, we then went to the robotics room and let them do their magic. They were pipetting reagents into our PCR plates and glucose solution. Then the robot transferred our solutions into a clear bottom plates for the plate reader.
Wednesday, July 12, 2017
Recovery Day
Today was more about the analysis of our data we got from our bacterial growth. Raphael walked us through on how to analyze our data correctly using Excel or Google Sheets. We used the number we got from the plate reader, put them in a table directly related to the table we were given to pipette the solutions into the PCR plate, and calculate averages. We did many calculations including charts of different samples. After all of the calculations, we made media containing a 50% Glycerol solution and our bacteria to cool it down without killing the bacteria.
Thursday, July 13, 2017
What’s on the Third Floor?
 Today was quite an interesting day. We got a tour of the third floor for the first time. We got a tour of the ABPDU (Advanced Biofuels Process Demonstration Unit) and got to see all the things that make the process of producing biofuels a little easier. We saw a lot of equipment: from centrifuges to equipment that separated the polysaccharides to monosaccharides and more. It was an interesting opportunity to learn that there are so many products that were developed over the years.
Today was quite an interesting day. We got a tour of the third floor for the first time. We got a tour of the ABPDU (Advanced Biofuels Process Demonstration Unit) and got to see all the things that make the process of producing biofuels a little easier. We saw a lot of equipment: from centrifuges to equipment that separated the polysaccharides to monosaccharides and more. It was an interesting opportunity to learn that there are so many products that were developed over the years.
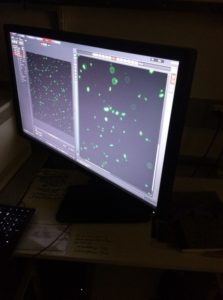
Friday, July 14, 2017
Bacterial Transformation
Today was definitely one of the coolest days. We platted our bacteria with the plasmas on media with LB and the corresponding antibiotic whether it was Green Fluorescent Protein or Red Fluorescent Protein that was added to the bacteria. After 1 hour of shaking in 37 degrees Celsius we then vortexed it and put it on slides to look under the microscope. What we saw was very cool. Now we get to say that we genetically modified E. Coli. How many people can say that?