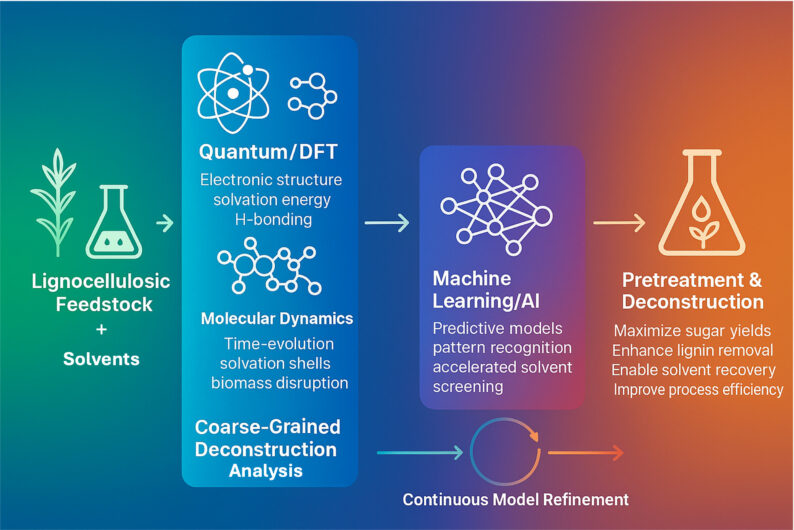
Background
The goals of the predictive deconstruction group are to 1) develop computational approaches to predicting the pretreatment efficacy of proposed new solvent systems and 2) generate molecular level understanding of solvent – biomass interactions via molecular mechanical and quantum calculation methods to better inform our predictive modeling methods. The landscape of potential solvents, including ionic liquids (IL), deep eutectic solvents (DES), and protic amines, available for biomass pretreatment is vast and, and when combined with potential process requirements such as solvent recovery and reuse, low toxicity to enzymes and microbial hosts, experimentally investigating the space of possibilities is inaccessible. The ability to predict the efficacy of a proposed new integrated solvent-based deconstruction process will dramatically expand the landscape of solvents that can be investigated for biomass pretreatment by conducting the bulk of the initial screening computationally. Predictive models also open the possibility to better engineer conversion technologies where models of monosaccharide and lignin fragment yield as a function of solvent, enzymes and pretreatment conditions are used to optimize pretreatment process conditions to maximize product yields. The predictive deconstruction group works very closely with the Biological Lignin Depolymerization, Catalytic Lignin Depolymerization, and Pretreatment Optimization and Process Integration groups and facilitated by the JBEI Feedstocks-to-Fuels Pipeline.
Research
Research in predictive deconstruction is aimed at developing fundamental understanding of the interactions among solvent systems (Amines, ILs, DESs, water) and the different components of lignocellulosic biomass that promote dissolution of cellulose, hemicellulose, and lignin. This information is being expanded and used to predict how new proposed solvents will perform under different pretreatment process scenarios. An additional focus is understanding how solvents interact with enzymes and other biological entities where it is important to understand and mitigate negative impacts of proposed solvents on enzyme activity and microbial growth and productivity. Ultimately, we would like to use the framework to design and optimize biomass and pretreatment process specific solvent systems.
Atomistic simulations of biomass-solvent systems
We use molecular dynamics simulations and quantum mechanics calculations such as COnductor like Screening MOdel for Real Solvents (COSMO-RS) to both explain how different solvents solubilize biomass and to the calculate properties (e.g., solubility parameters, ionic and electrostatic interactions, order parameters) of solvent – biomass mixtures that might be correlated with efficient biomass deconstruction. Insights and features extracted from these simulations are being used to generate machine learning models of pretreatment efficacy and biocompatibility.
Machine learning models
Machine learning models of biomass deconstruction using IL, DES and amine solvent systems are being constructed using features of the solvent extracted from atomistic simulations, from biomass characterization (e.g., composition, FT-IR, NMR), from the chemical structures of solvents, and from metadata on experimental conditions. To increase the effectiveness and design of the pretreatment process, machine learning techniques were also used to forecast the thermophysical characteristics of various solvent classes. Hypotheses such as whether a particular solvent system is agnostic to the feedstock phenotype will be tested by comparing models generated using different subsets of features (e.g., models including feedstock phenotype differences vs models excluding biomass phenotype differences). Predictive models are validated using JBEI’s feedstock to fuels pipeline, which is also be used to generate data designed to improve existing models.
Featured Publications
- Enzymatic cleavage of model lignin dimers depends on pH, enzyme, and body type. Sci. Rep. (2025)
- Volatile fatty acid extraction from fermentation broth using a hydrophobic ionic liquid and in situ enzymatic esterification. RSC Sustainability. (2025)
- Multi-scale computational screening and mechanistic insights of cyclic amines as solvents for improved lignocellulosic biomass processing. RSC Sustainability. (2025)
- Computational Advances in Ionic Liquid Applications for Green Chemistry: A Critical Review of Lignin Processing and Machine Learning Approaches. Molecules. (2024)
- Volatile fatty acid extraction from fermentation broth using a hydrophobic ionic liquid and in situ enzymatic esterification. RSC Sustainability. (2024)
- An engineered laccase from Fomitiporia mediterranea accelerates lignocellulose degradation. Biomolecules. (2024)
- Multiscale molecular simulations for the solvation of lignin in ionic liquids. Sci. Rep. (2023)
- Prediction of Solubility Parameters of Lignin and Ionic Liquids Using Multi-resolution Simulation Approaches. Green Chem. (2022)
- Effect of cosolvent on the solubility of glucose in ionic liquids: experimental and molecular dynamics simulations. Fluid Phase Equilib. (2022)
- Multiscale molecular simulation strategies for understanding the delignification mechanism of biomass in cyrene. ACS Sustain. Chem. Eng. (2022)
- Towards understanding of delignification of grassy and woody biomass in cholinium-based ionic liquids. Green Chem. (2021)
- A predictive toolset for the identification of effective lignocellulosic pretreatment solvents: a case study of solvents tailored for lignin extraction. Green Chem. (2021)
- Theoretical study on the microscopic mechanism of lignin solubilization in Keggin-type polyoxometalate ionic liquids. Phys. Chem. Chem. (2020)
- Theoretical Insights into the Role of Water in the Dissolution of Cellulose Using IL/Water Mixed Solvent Systems. J. Phys. Chem. (2015)
- Understanding pretreatment efficacy of four cholinium and imidazolium ionic liquids by chemistry and computation. Green Chem. (2014)
- Simulations reveal conformational changes of methylhydroxyl groups during dissolution of cellulose Iβ in ionic liquid 1-ethyl-3-methylimidazolium acetate. J. Phys. Chem. (2012)
- Molecular dynamics study of polysaccharides in binary solvent mixtures of an ionic liquid and water. J. Phys. Chem. (2011)
- Understanding the interactions of cellulose with ionic liquids: a molecular dynamics study. J. Phys. Chem. (2010)
Ionic Liquid Databases
- Public Literature Review: JBEI Pretreatment Lignin Ionic Liquids
- ILThermo
- The Ionic Liquid Property Explorer
- Reference https://www.mdpi.com/2306-5729/4/2/88
- Enterprise Ionic Liquids Database (ILUAM)