-By Irina Silva
JBEI develops new synthetic biology tools for engineering Saccharomyces cerevisiae
Pioneering work has been led by the U.S. Department of Energy’s Joint BioEnergy Institute (JBEI) to engineer microbes to transform plant derived starting materials into energy-rich biofuels. But despite the progress in genomics and synthetic biology for the optimization of biofuel production in engineered microbes, microbial engineering methods remain slow and laborious. Such is the case of the fungal host, Saccharomyces cerevisiae. The yeast S. cerevisiae has proven to be an excellent organism for commercial-scale production of biological molecules, though its strain development remains painstakingly slow due to difficulties related to the combined effect of different expression parts and host conditions. Now, researchers at JBEI have developed the largest, most comprehensive Cas9-based toolkit to quickly institute genetic changes in S. cerevisiae to optimize heterologous gene expression.
“S. cerevisiae is a promising microbial host for biofuel production. It is generally regarded as safe (GRAS), and has capacity for high-density fermentation. To build on our findings at JBEI we have developed new tools to enable more sophisticated strain engineering,” said Aindrila Mukhopadhyay, vice president for fuels synthesis at JBEI and the senior author of the study. “Our toolkit is available online to the scientific community and can be applied to other applications beyond biofuel privation in yeast.”
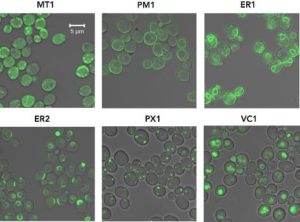
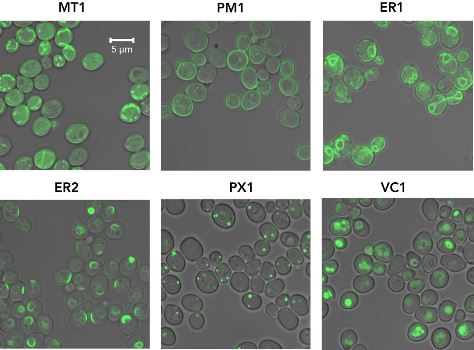
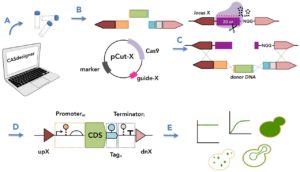
“CRISPR/Cas9 technology was employed to build a cloning-free toolkit that addresses commonly encountered obstacles in metabolic engineering, including chromosomal integration locus and promoter selection, as well as protein localization and solubility,” explained Amanda Reider Apel, Post-Doctoral Researcher at JBEI and co-first author of the study along with Leo d’Espaux and Maren Wehrs. “We constructed high-efficiency, Cas9-sgRNA plasmids targeting 23 characterized integration loci, characterized 37 standardized promoters in different growth phases and media, and validated the use of 10 protein tags conferring specific protein localization, turnover or solubility.”
The applicability of the toolkit was demonstrated by optimizing the expression of a challenging but industrially important enzyme, taxadiene synthase (TXS). This approach enabled the team to diagnose an issue with TXS solubility, the resolution of which yielded a 25-fold improvement in taxadiene production, the highest levels reported to date.
The strains and DNA sequences reported in the study published by Nucleic Acids Research have been deposited in the JBEI registry. The manuscript describing the toolkit and its application entitled, “A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae” was authored by Amanda Reider Apel, Leo d’Espaux, Maren Wehrs, Daniel Sachs, Rachel A. Li, Gary J. Tong, Megan Garber, Oge Nnadi, William Zhuang, Nathan J. Hillson, Jay D. Keasling and Aindrila Mukhopadhyay.