JBEI researchers define a systems-level model for cellular respiration
-By Irina Silva
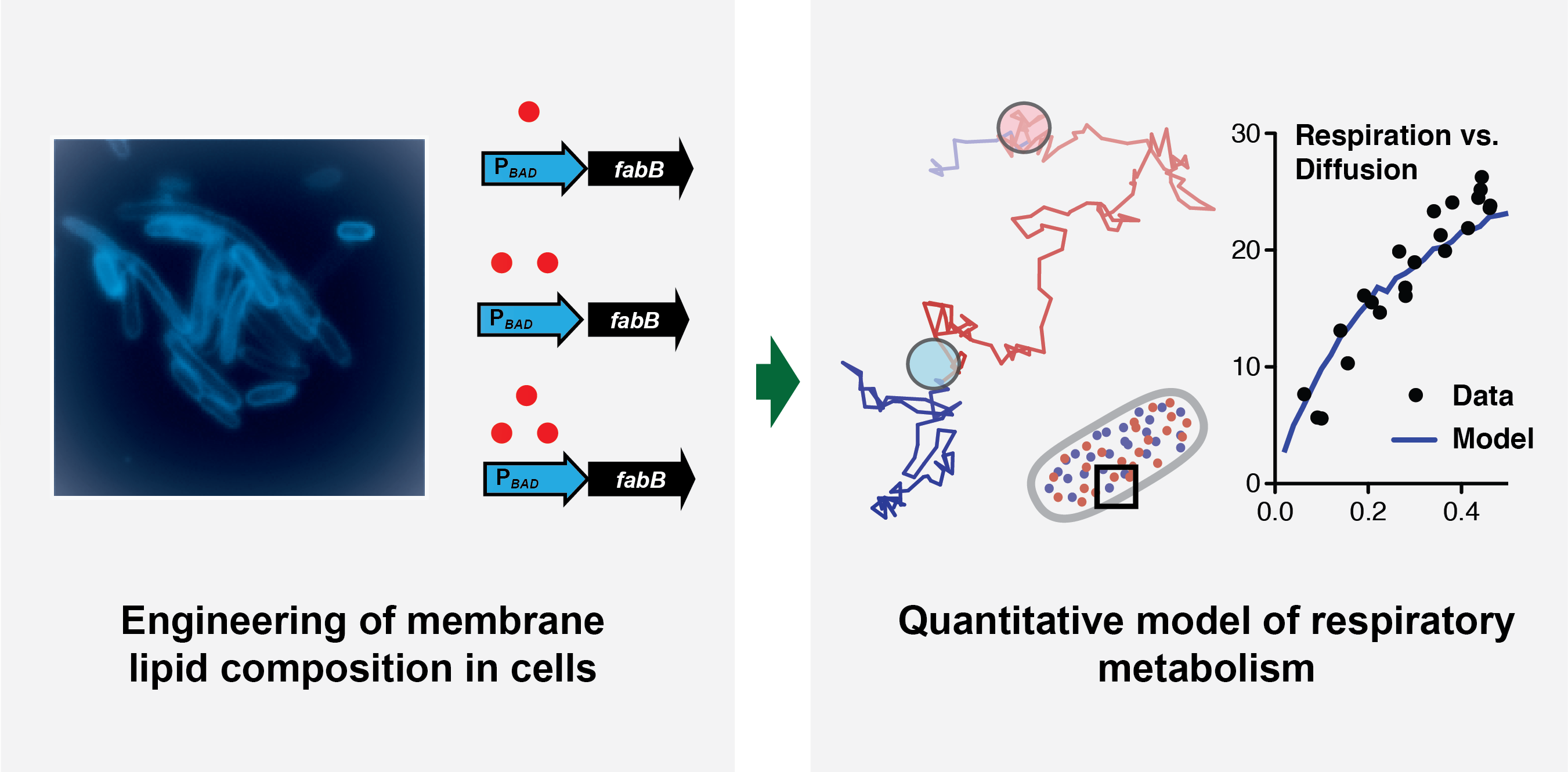
While much is known about how enzymes and molecules are involved in cellular respiration, the understanding of the respiration system as a whole remains limited. Researchers at the Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI) have gained insight into how cellular respiration is affected by the membrane environment in which it occurs. By engineering lipid synthesis to carefully control the membrane composition, researchers found that lipids, which consist of fatty acid molecules and determine membrane viscosity, also tightly control the rate of bacterial and yeast respiration. As lipid synthesis in these hosts is often engineered in order to produce molecules, these findings suggest new ways by which the pathways to produce biofuels and bioproducts could be optimized to maintain proper respiratory function, thereby increasing production.
This work was described in a paper, “Viscous control of cellular respiration by membrane lipid composition” which was published in Science on October 25. The research team was led by JBEI’s Chief Executive Officer Jay Keasling, corresponding author and also senior faculty scientist at Lawrence Berkeley National Laboratory. In this study, the researchers determined the relationship of membrane viscosity to cellular respiration. Viscosity, in the context of this paper, refers to how fluid a membrane is, and can fluctuate depending on which fatty acids are present.
“We were inspired by an old observation that cells have feedback mechanisms that allow their membrane structure to change”, said Itay Budin, JBEI researcher and lead author of the publication. “So we asked ourselves: How will changes in the types of lipids produced by bacterial cells affect their growth and metabolism?” To find an answer, Budin used synthetic biology and metabolic engineering methods to manipulate lipid synthesis and thereby carefully control membrane composition.
Lipids are commonly known as fats and oils. The split into these two categories depends on the physical state of the different types of fatty acids molecules at room temperature. For example, there are small chemical differences (double bonds) in the chemical structures of lipid molecules in solid butter and liquid olive oil, and these are responsible for their characteristic physical properties. Similarly, the physical properties of cell membranes, oily structures only a few nanometers thick, is also dependent on the chemistry of their lipid components. In this paper, the team modulated the viscosity of membranes in cells using metabolic engineering, a process by which JBEI researchers regularly use to control the relative levels of different chemical pathways in cells.
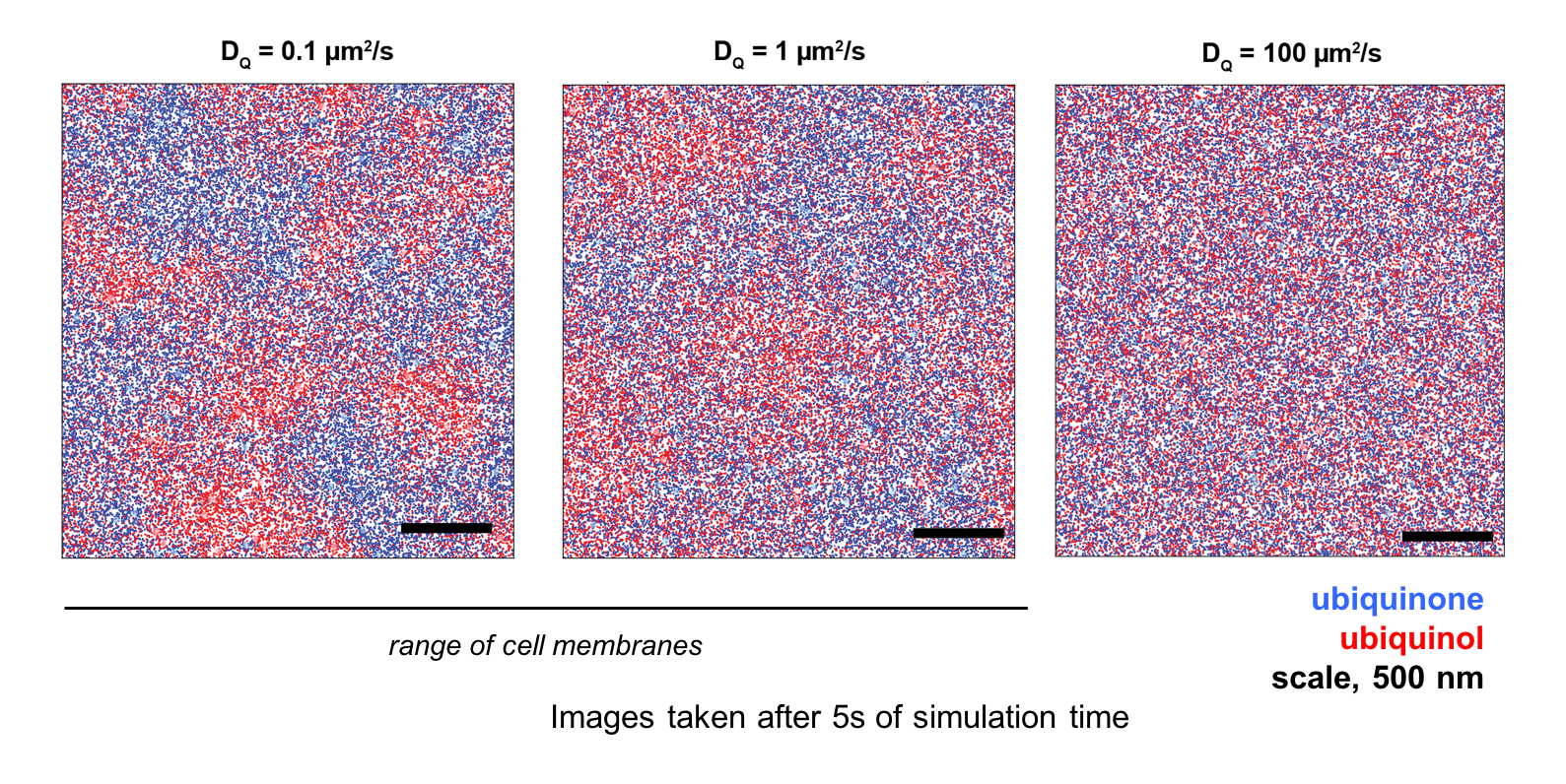
Budin found that lipids that determine membrane viscosity also tightly control the rate at which bacteria carried out respiration. Cellular respiration occurs through a set of reactions that occur when different enzymes and their substrates collide in the membrane, and viscosity sets the rate at which these collisions occur by random thermal motion (diffusion). Because of this key insight, Budin, working alongside former JBEI graduate student Tristan de Rond, developed a mathematical model for respiration that accounts for the diffusion of its components within the membrane. As inputs, they used quantitative measurements of the abundance and diffusion of the molecular components in the process, which was aided by mass spectrometry work by co-authors Yan Chen, Leanne Jade G. Chan, and Christopher J. Petzold. The team’s model described several aspects of bacterial metabolism, such as how it responds to inhibitors or changes in enzyme concentrations. They then showed that lipids also mediate respiratory rates in mitochondria, dedicated organelles used by all eukaryotic cells for energy production. Thus, lipids could effectively set the ‘speed limit’ by which cells can ‘breathe’ through their effects on membrane diffusion.
“Itay’s research provides us a better understanding of the central metabolism in the two most commonly used hosts used for biotechnology: E. coli and S. cerevisiae,” said Keasling. “This is knowledge with ample application in future metabolic engineering efforts. Furthermore, it demonstrates how tools developed by synthetic biology can also be applied to address fundamental questions in biology.”
JBEI is a DOE Bioenergy Research Center funded by DOE’s Office of Science, and is dedicated to establishing the scientific knowledge and new technologies to transform the maximum amount of carbon available in bioenergy crops into biofuels and bioproducts. This work was also supported by funding from the National Science Foundation.
