Research Goal
To lead the continuous development and maintenance of SynBio software and laboratory automation infrastructure, and operation of DIVA services.
Projects
- ICE Repository of microbial strains, (DNA and protein) sequences, and plant seeds/lines
- Bioparts.org Search portal across repositories of strains and sequences (ICE and beyond)
- DIVA DNA Design, Implementation, Validation, Automation platform and services
- EDD Experiment Data Depot
Featured Media
My Perfect Path podcast (S2/Ep6) “Adapting fast is important for your career. Fail Smarter. Succeed Sooner”
First Author Conversations Podcast. Episode 3: Application of Synthetic Biology
Understanding and Designing Flanking Homology DNA Assembly Experiments
Nathan Hillson talks with the Risk Group about the: “BioEconomy: Promise and Perils of Synthetic Biology”
Nathan Hillson talks at National Academics Biotech Study Second Public Meeting
Session 2: Enabling Tools
Session 6: Tools Enhancing Risk Analysis
Nathan Hillson talks about the role of software development at JBEI
Microfluidic Device Mixes And Matches DNA For Synthetic Biology
Cold Spring Harbor Laboratory Seminar: Synthetic Biology Scalability and Responsible Innovation

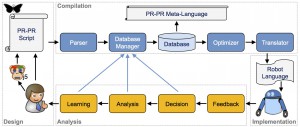
Training Your Robot the PR-PR Way
Latest JBEI Startup to Speed Up Biotech Industry
DNA Construction Software Saves Time, Resources and Money
Featured Publications
- “High-Throughput Microfluidic Electroporation (HTME): A Scalable, 384-Well Platform for Multiplexed Cell Engineering” Bioengineering (2025)
- “The tier system: a host development framework for bioengineering” Current Opinion in Biotechnology (2025)
- “Machine learning-led semi-automated medium optimization reveals salt as key for flaviolin production in Pseudomonas putida” Commun Biol (2025)
- “Binary vector copy number engineering improves Agrobacterium-mediated transformation“. Nat Biotechnol (2024)
- “Bioinformatic prediction and high throughput in vivo screening to identify cis-regulatory elements for the development of algal synthetic promoters” ACS Synthetic Biology (2024)
- “A Biomanufacturing Plan to Confront Future Biological Threats: Expert Panel Policy Review and Near-Term Recommendations” Council on Strategic Risks (2024)
- “Automated Platform for the Plasmid Construction Process” ACS Synthetic Biology (2023)
- “Alkaline-SDS cell lysis of microbes with acetone protein precipitation for proteomic sample preparation in 96-well plate format” PLOS ONE (2023)
- “Ten simple rules for managing laboratory information.” PLoS computational biology (2023)
- “A Procedural Framework for Benchmarking Biofoundry Capabilities“ ACS Synthetic Biology (2023)
- “Introduction to the Special Issue on BioFoundries and Cloud Laboratories” J. Emerg. Technol. Comput. Syst (2023)
- “Artificial Intelligence and Machine Learning for Bioenergy Research: Opportunities and
Challenges” U.S. Department of Energy Office of Science and Office of Energy Efficiency and Renewable Energy (2023) - “Biorisk Management Case Study: United States Department of Energy Joint Genome Institute” Stanford Digital Repository (2023)
- “Perspectives for self-driving labs in synthetic biology” Current Opinion in Biotechnology (2023)
- “Scalable and automated CRISPR-based strain engineering using droplet microfluidics” Microsyst Nanoeng (2022)
- “Modular automated bottom-up proteomic sample preparation for high-throughput Applications”. PLoS ONE (2022)
- “Making Security Viral: Shifting Engineering Biology Culture and Publishing”. ACS Synthetic Biology (2022)
- “BioParts—A Biological Parts Search Portal and Updates to the ICE Parts Registry Software Platform” ACS Synthetic Biology (2021)
- “Integration of Proteomics and Metabolomics Into the Design, Build, Test, Learn Cycle to Improve 3-Hydroxypropionic Acid Production in Aspergillus pseudoterreus” Frontiers in Bioengineering and Biotechnology (2021)
- “Multiomics Data Collection, Visualization, and Utilization for Guiding Metabolic Engineering“. Frontiers in Bioengineering and Biotechnology (2021)
- “Machine learning for metabolic engineering: A review” Metabolic Engineering (2020)
- “Repurposing a microfluidic formulation device for automated DNA construction” PLOS One (2020)
- “An Integrated Computer-Aided Design and Manufacturing Workflow for Synthetic Biology” DNA Cloning and Assembly (2020)
- “Combinatorial-Hierarchical DNA Library Design Using the TeselaGen DESIGN Module with j5” DNA Cloning and Assembly (2020)
- “DNA Scanner: a web application for comparing DNA synthesis feasibility, price, and turnaround time across vendors” OUP Synthetic Biology (2020)
- “Embrace experimentation in biosecurity governance” Science (2020)
- “Adenosine Triphosphate and Carbon Efficient Route to Second Generation Biofuel Isopentanol” ACS Synthetic Biology (2020)
- “Fluorescent amplification for next generation sequencing (FA-NGS) library preparation” BMC Genomics (2020)
- “Enhancing Terminal Deoxynucleotidyl Transferase Activity on Substrates with 3′ Terminal Structures for Enzymatic De Novo DNA Synthesis” Genes (2020)
- “Mevalonate Pathway Promiscuity Enables Noncanonical Terpene Production” ACS Synthetic Biology (2019)
- “An automated ‘cells-to-peptides’ sample preparation workflow for high-throughput, quantitative proteomic assays of microbes” J Proteome Res (2019)
- “Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases” J Ind Microbiol Biotechnol (2019)
- “Lessons from Two Design–Build–Test–Learn Cycles of Dodecanol Production in Escherichia coli Aided by Machine Learning” ACS Synthetic Biology (2019)
- “New Structural insights into dehydratase substrate selection for the borrelidin and fluvirucin polyketide synthases” JIMB (2019)
- “Building a global alliance of biofoundries“, Nature Communications (2019)
- “Parallel Integration and Chromosomal Expansion of Metabolic Pathways“, ACS Synthetic Biology (2018)
- “Jungle Express is a versatile repressor system for tight transcriptional control“, Nature Communications (2018)
- “De novo DNA synthesis using polymerase-nucleotide conjugates“, Nature BioTech (2018)
- “Biochemical Characterization of β-Amino Acid Incorporation in Fluvirucin B2 Biosynthesis“, ChemBioChem (2018)
- “Isolation and characterization of novel mutations in the pSC101 origin that increase copy number“, Scientific Reports (2018)
- “A combinatorial approach to synthetic transcription factor-promoter combinations for yeast strain engineering” Yeast (2018)
- “Oxidative cyclization of prodigiosin by an alkylglycerol monooxygenase-like enzyme”, Nature Chemical Biology (2017)
- “The Experiment Data Depot: a web-based software tool for biological experimental data storage, sharing, and visualization”, ACS Synthetic Biology (2017)
- “Droplet Microfluidics for Synthetic Biology”, Lab on a Chip (2017)
- “Streamlining the Design-to-Build transition with Build-Optimization Software Tools (BOOST)”, ACS Synthetic Biology (2016)
- “A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae”, Nucleic Acids Research (2016).
- “Develop an improved metabolic engineering method for modifying microorganisms for biofuel production from cellulosic sugars”, JBEI end-of-year report (2016)
- “New tools available for facilitating the design of engineered pathways in microorganisms for biofuel production from cellulosic sugars”, JBEI quarterly report (2016)
- “Improving Synthetic Biology Communication: Recommended Practices for Visual Depiction and Digital Submission of Genetic Designs”, ACS Synthetic Biology (2016)
- “Investigation of Proposed Ladderane Biosynthetic Genes from Anammox Bacteria by Heterologous Expression in E. coli“, PLOS ONE (2016)
- “End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis”, J Biol Eng (2016)
- “A Droplet Microfluidic Platform for Automating Genetic Engineering”, ACS Synthetic Biology (2016)
- “SBOL Visual: A Graphical Language for Genetic Designs”, PLOS Biology (2015)
- “A Versatile Microfluidic Device for Automating Synthetic Biology”, ACS Synthetic Biology (2015)
- “Industrialization of Biology: A Roadmap to Accelerate the Advanced Manufacturing of Chemicals“, The National Academies Press (2015)
- “The Joint Genome Institute’s synthetic biology internal review process”, Journal of Responsible Innovation (2015)
- “Principal component analysis of proteomics (PCAP) as a tool to direct metabolic engineering”, Metabolic Engineering (2014)
- “Characterization of Wastewater Treatment Plant Microbial Communities and the Effects of Carbon Sources on Diversity in Laboratory Models”, PLOS ONE. (2014)
- “The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology”, Nat Biotechnol. (2014)
- “The Plant Glycosyltransferase Clone Collection for Functional Genomics”, Plant J. (2014)
- “Cloud-Enabled Microscopy and Droplet Microfluidic Platform for Specific Detection of Escherichia coli in Water”, PLoS ONE (2014).
- “PR-PR Cross-Platform Laboratory Automation System”, ACS Synthetic Biology (2014)
- “j5 DNA Assembly Design Automation”, DNA Cloning and Assembly Methods (2014)
- “Development of a broad-host synthetic biology toolbox for ralstonia eutropha and its application to engineering hydrocarbon biofuel production”, Microbial Cell Factories (2013)
- ” Proposed design of distributed macroalgal biorefineries: thermodynamics, bioconversion technology, and sustainability implications for developing economies”, Biofuels, Bioprod. Bioref. (2013)
- “Engineering of Ralstonia eutropha H16 for Autotrophic and Heterotrophic Production of Methyl Ketones”, Appl. Environ. Microbiol. (2013)
- “The Fusion of Biology, Computer Science, and Engineering – towards efficient and successful synthetic biology”, Perspectives in Biology and Medicine (2013)
- “Functionalizing bacterial cell surfaces with a phage protein”, Chemical Communications (2013)
- “Distributed marine biorefineries for developing economies”, Proceedings of the ASME 2012 International Mechanical Engineering Congress & Exposition (2012)
- “PaR-PaR Laboratory Automation platform”, ACS Synthetic Biology (2012).
- “Design, Implementation and Practice of JBEI-ICE: An Open Source Biological Part Registry Platform and Tools”, Nucleic Acids Research (2012)
- “DeviceEditor visual biological CAD canvas”, Journal of Biological Engineering (2012).
- “j5 DNA Assembly Design Automation Software”, ACS Synthetic Biology (2012)
- “Microbial stress tolerance: from genomics to biofuels”, Microbial Stress Tolerance for Biofuels (2012)
- “DNA Assembly Method Standardization for Synthetic Biomolecular Circuits and Systems”, Design and Analysis of Bio-molecular Circuits (2011)
Featured Intellectual Property
- Scar-Less Multi-Part DNA Assembly Design Automation. (2019) U.S. Patent 11,749,379
- Scar-Less Multi-Part DNA Assembly Design Automation. (2016) U.S. Patent 10,373,703
- Scar-Less Multi-Part DNA Assembly Design Automation. (2012) U.S. Patent 9,361,427
- Microfluidic Platform for Synthetic Biology Applications. (2012) U.S. Patent Application US20120258487
- Heavy Metal Biosensor. (2008) U.S. Patent Application US20110117590