JBEI researchers pave the way for efficient gene expression at any scale
-By Lida Gifford
In the quest to find the key to a rainforest dwelling bacterium’s lignin-degrading ability, researchers at the Department of Energy (DOE)’s Joint BioEnergy Institute (JBEI) have constructed a gene expression system that outperforms conventional systems. Controlling gene expression is crucial to scientists’ ability to perform basic science and biotechnological research to produce enzymes, bio-based products, and biofuels, both at the bench and on industrial scales.
The JBEI team was led by Michael Thelen, a biochemist in the Deconstruction Division, and included researchers from Lawrence Berkeley National Laboratory (Berkeley Lab), Lawrence Livermore National Laboratory (LLNL), and San Francisco State University. Their work, published on September 6 in Nature Communications, describes the bottom-up engineering of Jungle Express, a versatile expression system that enables efficient gene regulation in diverse gram-negative bacteria.
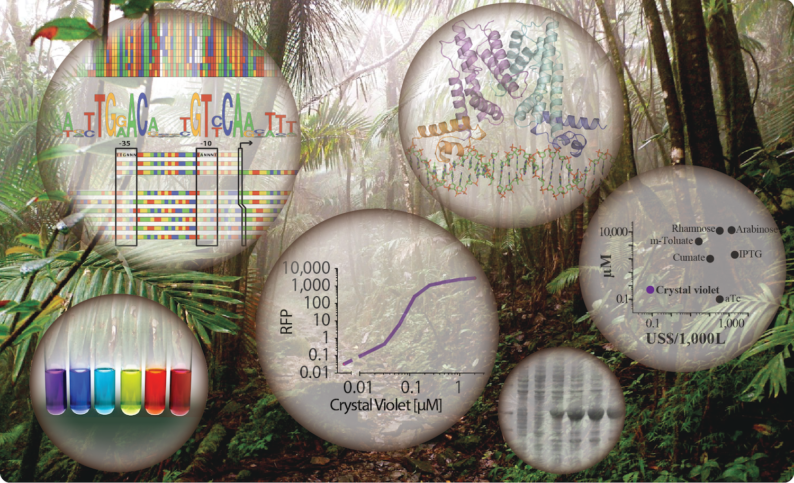
Thomas Ruegg, JBEI researcher and lead author of the publication, said that he began developing this system while studying Enterobacter lignolyticus, a soil bacterium native to a tropical rainforest in Puerto Rico, giving rise the name Jungle Express. Two genes in E. lignolyticus allow the bacterium to withstand exposure to harsh ionic liquids that are used in the deconstruction of biomass, a necessary step in the production of biofuels. Ruegg focused on the regulatory component of the resistance mechanism and tested its response to a range of chemicals that share certain properties with ionic liquids. One of those chemicals was crystal violet, an antifungal agent commonly found in microbiology labs that is also used as a dye for textiles and printing inks. “When I saw extremely high sensitivity to crystal violet,” said Ruegg, “I decided to engineer a gene expression system that can be efficiently activated by this cheap and readily available resource.”

The researchers performed a combination of computational analysis and rational molecular engineering approaches to develop, understand and optimize performance of Jungle Express. This system encompasses several qualities that are very desirable in gene expression applications: tight control, high level and specificity of gene expression, versatility of host bacteria (from E. coli to industrially relevant strains), cost-effectiveness, and flexibility.
To further characterize the system at the molecular level, Jose Henrique Pereira, a research scientist in JBEI’s Technology Division, performed X-ray crystallography at the Advanced Light Source, a DOE Office of Science User Facility. Using these data, they determined the interactions between the regulatory elements and two molecules, including crystal violet, used to turn on the system, which gives insight into its specificity.
“Our findings have the potential to overcome the bottlenecks encountered in earlier systems, and open the way for tightly controlled and efficient gene expression that is not restricted to host organism, substrate, or scale,” explained Thelen, who is also a biochemist at LLNL. “Overall, this has been a fascinating journey that literally started in a jungle of microbial genetic information,” said Ruegg. “We explored this tremendous resource and were able to change the context for the development of a novel game-changing application.”
JBEI is a DOE Bioenergy Research Center funded by DOE’s Office of Science, and is dedicated to developing advanced biofuels. Other co-authors on the paper are: Joseph Chen, Andy DeGiovanni, Giovanni Tomaleri, Steve Singer, Nathan Hillson, Blake Simmons, and Paul Adams of JBEI and Pavel Novichkov and Vivek Mutalik of the Environmental Genomics and Systems Biology Division at Berkeley Lab. Read more about this research in the LLNL press release.