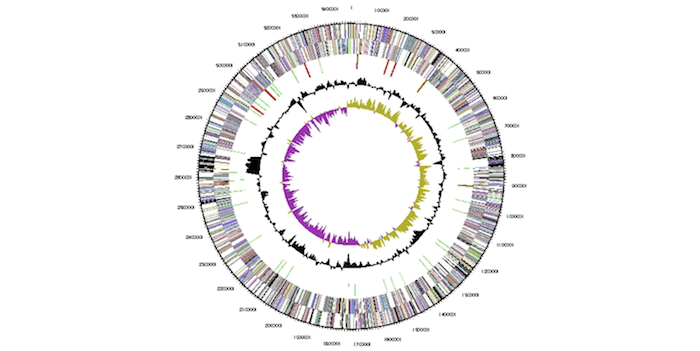
Multiple annotated DNA sequences, DNA design and assembly files, primers, plasmids and strains have been produced, validated, and shared. Explore these and microarray datasets for yeast responses to biofuel-related stress. Image from Chertkov O, et al., Stand. Genomic Sci. 5:1 (2011).
Biofuels Pathways Proteomic and 16s iTag data
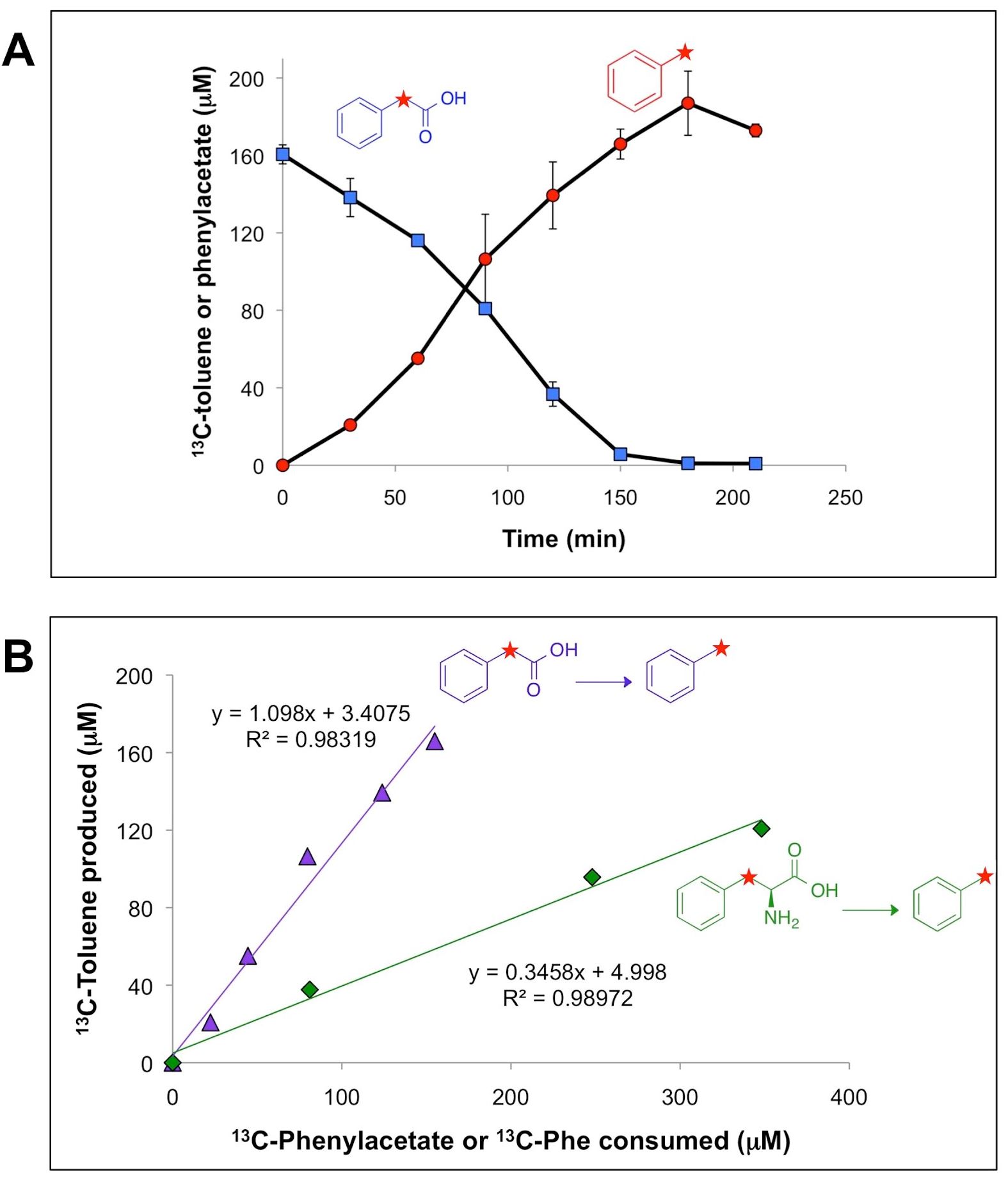
Researchers at JBEI have investigated an enzyme that could enable first-time biochemical production of the widely used octane booster, toluene (Zargar et al. 2016). For this work, in vivo, in vitro, metagenomic, and metaproteomic studies were conducted with an anaerobic microbial community derived from sewage sludge that quantitatively produced toluene from phenylacetic acid (via decarboxylation). Data included community composition by 16S rRNA gene iTag analysis and metaproteomic analysis of partially purified fractions of the community proteome that catalyzed toluene biosynthesis from phenylacetic acid; proteins were identified by mapping peptides to a community metagenome (Joint Genome Institute IMG Taxon ID 3300001784).
Host Engineering Microarray Data
The yeast Saccharomyces cerevisiae can be used to convert biomass to biofuels. In order to better understand yeast response due to stressors that could inhibit or limit this process, yeast was evaluated under various conditions. Microarray analysis was performed at different time points to determine yeast genome-wide response when exposed to salt, acid, hypoxic conditions, and hydrogen peroxide, as well as ethanol, isopentenol, and other biofuels candidates.
Synthetic Biology Combinatorial Plasmids
PR-PR is an open-source biology friendly programming language that has utility as a cross-platform lab automation system. PR-PR can be used with liquid-handling robots, microfluidics, and microscopy, as well as for protocol translation into human languages. In the course of further developing PR-PR, researchers wrote protocols for processes to manipulate DNA: combinatorial modified Golden Gate DNA assembly, Kunkel DNA mutagenesis and hierarchical Gibson DNA assembly (Linshiz, et al., 2014). Information for initial and resulting materials is available in the JBEI registry*, including DNA sequences, annotated sequence and sequencing trace validation files, and design files.
Synthetic Biology Bacterial Strains & Plasmids
ScanDrop is an all-in-one microfluidic system for detecting and reporting the presence of bacteria in drinking water (Golberg, et al., 2014). Target bacteria were captured using antibody-coated magnetic beads and then encapsulated in picoliter-sized droplets with fluorescently labeled antibodies for detection. Plasmids expressing either red or green fluorescent proteins were transformed into a strain of E. coli and used to demonstrate the feasibility of this system. Specific information about these materials can be found in the public instance of the JBEI Registry*.
Synthetic Biology Bacterial Strains & Plasmids
Ralstonia eutropha is a bacterium that can grow using hydrogen and carbon dioxide in an aerobic environment. When food sources are limited, R. eutropha produces the biopolymer, polyhydroxybutyrate (PHB). A plasmid-based toolbox was developed to optimize and diversify production of hydrocarbons in these bacteria (Bi, et al., 2013). Users can log on to the public instance of the JBEI Registry* to view all 60 bacterial strains and plasmids, which contain a variety of origins of replication, promoters, and ribosomal binding sites.
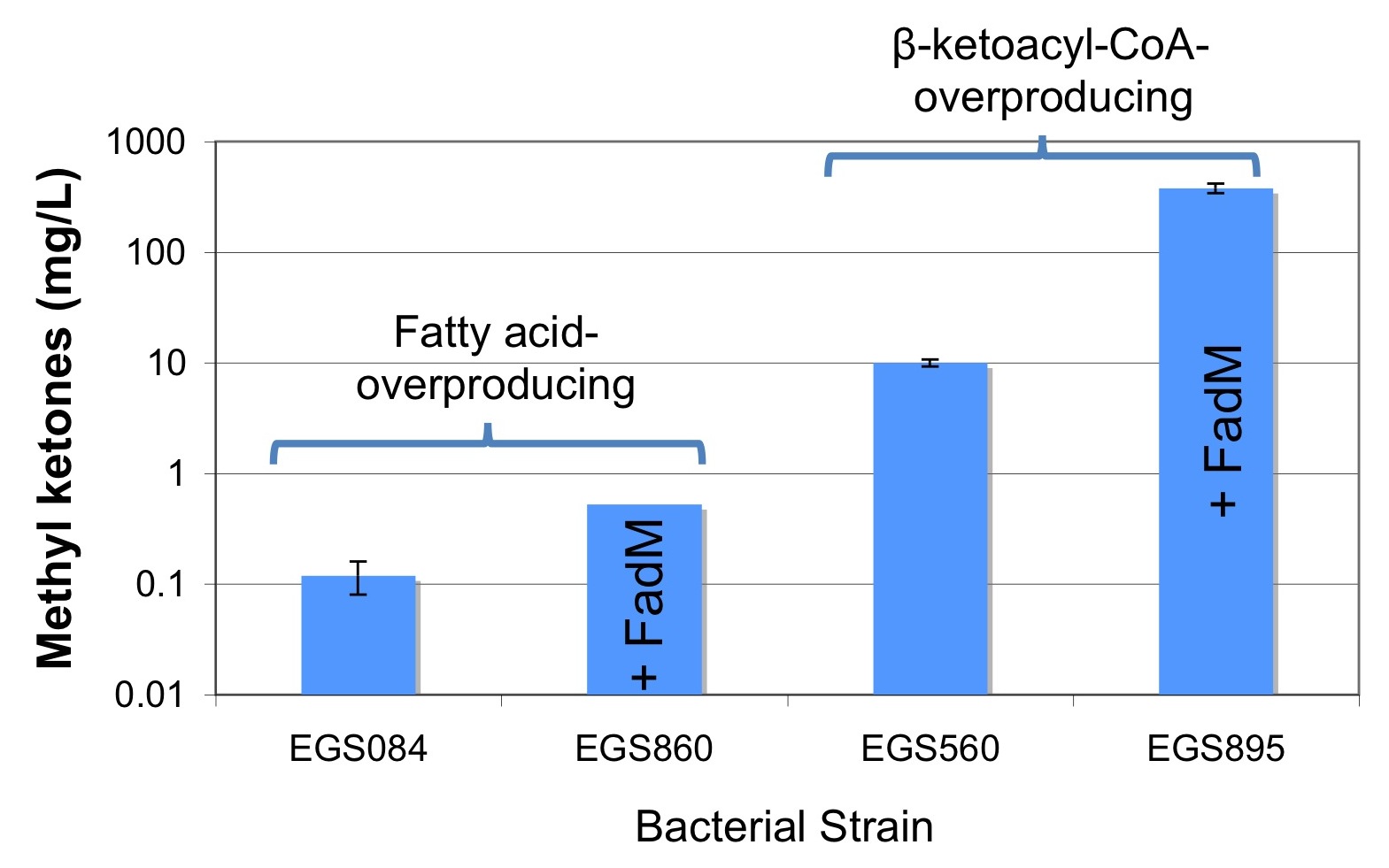
Biofuels Pathways Microarray Data
JBEI has engineered a new pathway for diesel-range methyl ketone biosynthesis in E. coli (Goh E-B, et al., 2012). Part of this effort involved discovery, through transcriptomic evidence (whole-genome microarrays), that up-regulation of the native fadM thioesterase in E. coli was correlated with enhanced methyl ketone titers.
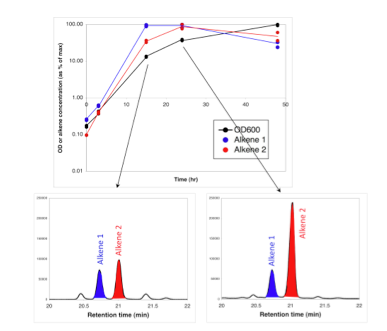
Biofuels Pathways Microarray Data
JBEI researchers identified three genes in the bacterium Micrococcus luteus that are associated with long-chain alkene biosynthesis from fatty acids (Beller H, et al., 2010). Related research (Pereira HJ, et al., 2012) used two different lines of evidence to explain the increase in the ratio of anteiso– to iso-branched alkenes that was observed during the transition from early to late stationary phase in M. luteus: structural studies of a key enzyme involved in fatty acid biosynthesis in M. luteus (FabH, or ß-ketoacyl-ACP synthase III) and transcriptional (whole-genome microarray) studies of M. luteus during different growth phases.
*Please note: Any user can create an account on the public instance of the JBEI Registry by going to https://public-registry.jbei.org/.